Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
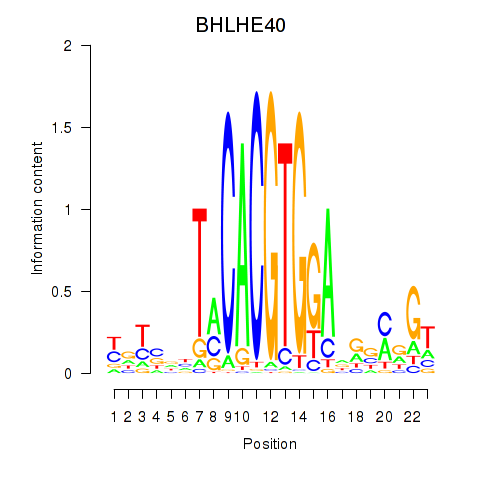
Results for BHLHE40
Z-value: 1.63
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg19_v2_chr3_+_5020801_5020952 | -0.88 | 3.3e-07 | Click! |
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.5 | 1.6 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.5 | 4.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 1.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.3 | 1.6 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 0.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 1.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.0 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.2 | 1.1 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 0.2 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.8 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.8 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.7 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 2.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.3 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 4.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.7 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.5 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 | 1.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 2.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.9 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 1.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 1.6 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.9 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 2.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 1.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 1.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 1.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 1.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.4 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.9 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.5 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.1 | 1.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 3.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 1.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.4 | 4.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 1.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.5 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 1.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.0 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 2.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 4.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |