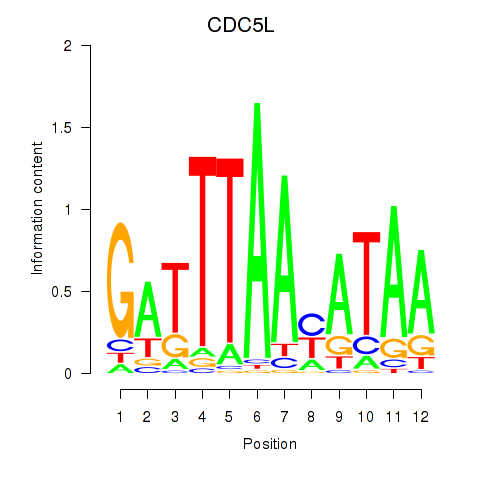
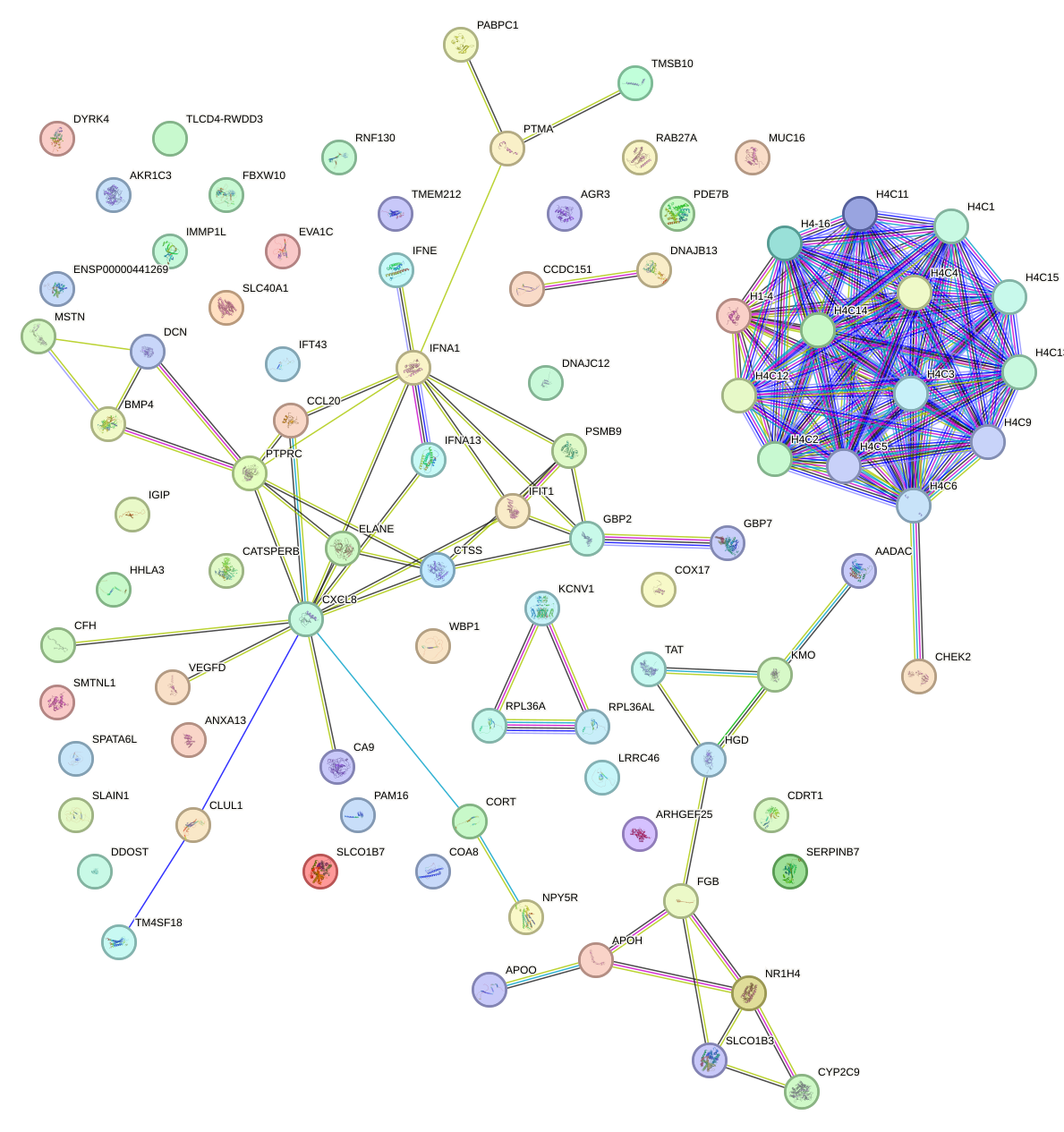
| 0.2 |
3.2 |
GO:0043152 |
induction of bacterial agglutination(GO:0043152) |
| 0.2 |
0.7 |
GO:0051097 |
negative regulation of helicase activity(GO:0051097) |
| 0.1 |
0.4 |
GO:0045362 |
regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 |
0.3 |
GO:1903522 |
regulation of blood circulation(GO:1903522) |
| 0.1 |
1.0 |
GO:0010898 |
positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 |
0.8 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.1 |
0.2 |
GO:1903988 |
spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 |
0.2 |
GO:0072428 |
signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 |
0.2 |
GO:2000224 |
sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 |
0.3 |
GO:2000006 |
apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 |
0.4 |
GO:1903435 |
positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 |
0.1 |
GO:0035378 |
carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 |
0.2 |
GO:2001248 |
nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 |
0.4 |
GO:0097056 |
selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 |
0.3 |
GO:1904158 |
axonemal central apparatus assembly(GO:1904158) |
| 0.0 |
0.0 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.0 |
0.6 |
GO:0051918 |
negative regulation of fibrinolysis(GO:0051918) |
| 0.0 |
0.3 |
GO:0016098 |
monoterpenoid metabolic process(GO:0016098) |
| 0.0 |
0.1 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 |
0.3 |
GO:0045415 |
negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 |
0.1 |
GO:2000502 |
negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 |
0.2 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.0 |
0.2 |
GO:0014050 |
negative regulation of glutamate secretion(GO:0014050) |
| 0.0 |
0.2 |
GO:0090362 |
positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 |
0.2 |
GO:1902725 |
negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 |
0.1 |
GO:0006864 |
pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 |
0.1 |
GO:0002879 |
leukocyte chemotaxis involved in inflammatory response(GO:0002232) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 |
0.2 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 |
0.3 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.0 |
0.1 |
GO:0055099 |
response to high density lipoprotein particle(GO:0055099) |
| 0.0 |
0.2 |
GO:1900063 |
regulation of peroxisome organization(GO:1900063) |
| 0.0 |
0.1 |
GO:1902283 |
negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 |
0.1 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.0 |
0.6 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.2 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.4 |
GO:0033141 |
positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 |
0.1 |
GO:1903348 |
positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 |
0.2 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 |
0.5 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.1 |
GO:0002528 |
regulation of vascular permeability involved in acute inflammatory response(GO:0002528) sarcomerogenesis(GO:0048769) |
| 0.0 |
0.1 |
GO:0006533 |
aspartate catabolic process(GO:0006533) |
| 0.0 |
0.3 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 |
0.1 |
GO:0031394 |
myoblast migration involved in skeletal muscle regeneration(GO:0014839) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 |
0.9 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.1 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.0 |
0.1 |
GO:0036023 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 |
0.1 |
GO:0060335 |
positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 |
0.1 |
GO:0046373 |
arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 |
0.1 |
GO:0015811 |
L-cystine transport(GO:0015811) |
| 0.0 |
0.1 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 |
0.1 |
GO:0031120 |
snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 |
0.1 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.0 |
0.2 |
GO:2000622 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.1 |
GO:2000435 |
regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 |
0.0 |
GO:0033693 |
neurofilament bundle assembly(GO:0033693) |
| 0.0 |
0.2 |
GO:1901078 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.1 |
GO:0009441 |
glycolate metabolic process(GO:0009441) |
| 0.0 |
0.1 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.0 |
GO:0002818 |
intracellular defense response(GO:0002818) |
| 0.0 |
0.1 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 |
0.1 |
GO:0045590 |
negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 |
0.1 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.1 |
GO:0009256 |
10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 |
0.2 |
GO:0070129 |
regulation of mitochondrial translation(GO:0070129) |
| 0.0 |
0.3 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.2 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.2 |
GO:1990573 |
potassium ion import across plasma membrane(GO:1990573) |
| 0.0 |
0.2 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.1 |
GO:0006021 |
inositol biosynthetic process(GO:0006021) |
| 0.0 |
0.1 |
GO:0090076 |
relaxation of skeletal muscle(GO:0090076) |
| 0.0 |
0.2 |
GO:0050713 |
negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 |
0.1 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.1 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 |
0.1 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.0 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 |
0.1 |
GO:2000111 |
primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 |
0.0 |
GO:0032072 |
plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |