Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
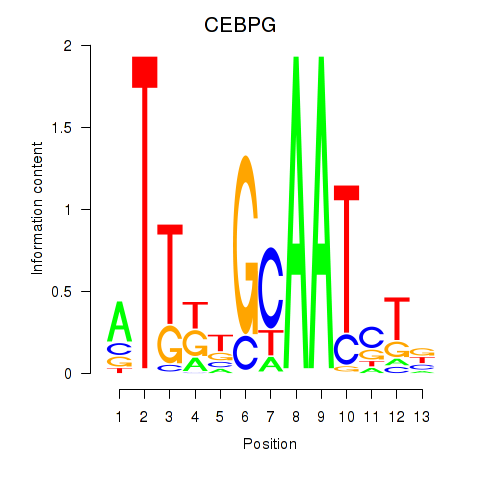
Results for CEBPG
Z-value: 1.99
Transcription factors associated with CEBPG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPG
|
ENSG00000153879.4 | CCAAT enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPG | hg19_v2_chr19_+_33865218_33865254 | -0.65 | 1.8e-03 | Click! |
Activity profile of CEBPG motif
Sorted Z-values of CEBPG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 11.8 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.8 | 3.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.7 | 2.2 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.5 | 2.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 1.5 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.5 | 3.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 1.3 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.4 | 7.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 0.7 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.3 | 12.2 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.3 | 2.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 | 0.8 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.2 | 6.6 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.2 | 1.6 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) cellular response to UV-C(GO:0071494) |
| 0.2 | 1.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.7 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 0.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 1.6 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 1.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.6 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.2 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.6 | GO:0045345 | positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 5.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 2.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 1.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 2.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.3 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 1.3 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 2.2 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.5 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0010841 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.9 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.6 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 1.8 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 1.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.4 | 2.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 16.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 0.6 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 14.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 13.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 3.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.3 | 1.0 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.3 | 16.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 0.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.3 | 3.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 1.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 2.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 0.6 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 4.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 6.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.5 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.4 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.2 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.1 | 12.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 2.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 10.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 2.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 6.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 7.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 6.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 2.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 10.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 3.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |