Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
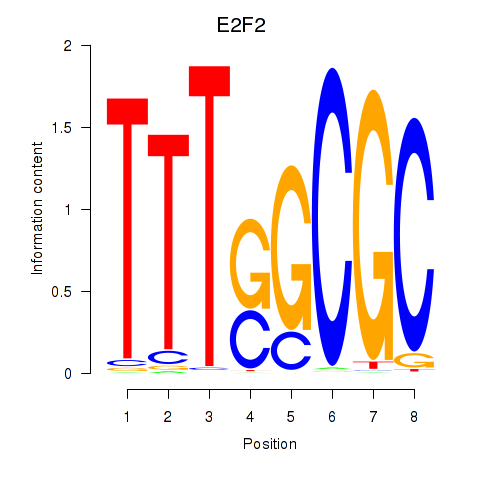
Results for E2F2_E2F5
Z-value: 8.58
Transcription factors associated with E2F2_E2F5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F2
|
ENSG00000007968.6 | E2F transcription factor 2 |
|
E2F5
|
ENSG00000133740.6 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F5 | hg19_v2_chr8_+_86089460_86089483 | 0.96 | 4.1e-11 | Click! |
| E2F2 | hg19_v2_chr1_-_23857698_23857733 | 0.92 | 7.7e-09 | Click! |
Activity profile of E2F2_E2F5 motif
Sorted Z-values of E2F2_E2F5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F2_E2F5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 39.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 5.8 | 23.3 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 5.5 | 71.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 5.4 | 5.4 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 5.4 | 16.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 5.1 | 20.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 5.0 | 35.0 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 4.4 | 30.8 | GO:0060356 | leucine import(GO:0060356) |
| 3.9 | 15.8 | GO:0006272 | leading strand elongation(GO:0006272) |
| 2.8 | 11.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 2.7 | 19.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 2.6 | 18.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 2.5 | 7.6 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 2.3 | 6.9 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 2.1 | 12.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 2.0 | 13.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 1.8 | 1.8 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.8 | 25.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.8 | 12.3 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 1.8 | 87.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.6 | 23.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.5 | 4.6 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 1.4 | 18.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.3 | 3.9 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 1.3 | 16.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 1.2 | 3.6 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 1.1 | 5.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.1 | 15.3 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 1.1 | 7.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 1.0 | 9.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 1.0 | 3.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 1.0 | 4.9 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 1.0 | 13.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.9 | 9.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.9 | 2.7 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.9 | 3.5 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.8 | 2.5 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.8 | 6.4 | GO:0033504 | floor plate development(GO:0033504) |
| 0.8 | 4.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.8 | 4.5 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.7 | 2.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.7 | 2.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.6 | 7.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 9.9 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.5 | 5.9 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.5 | 17.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 7.4 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.5 | 1.9 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.5 | 0.5 | GO:0035822 | gene conversion(GO:0035822) |
| 0.4 | 5.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.4 | 2.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 | 1.2 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.4 | 8.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 8.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 3.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 4.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.3 | 3.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 1.7 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.3 | 2.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.3 | 18.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.3 | 1.6 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.3 | 1.6 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.3 | 1.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.3 | 2.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.3 | 3.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.3 | 4.8 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.3 | 4.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.3 | 0.8 | GO:0060437 | lung growth(GO:0060437) |
| 0.3 | 2.5 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) |
| 0.3 | 5.9 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.3 | 3.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 | 1.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.3 | 2.4 | GO:0042148 | strand invasion(GO:0042148) |
| 0.2 | 4.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 2.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.2 | 1.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 1.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.2 | 18.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.2 | 6.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 3.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 19.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 7.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.2 | 0.6 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.2 | 11.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 4.6 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.2 | 2.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 1.5 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 1.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 17.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.2 | 21.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.7 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 1.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 3.7 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.2 | 2.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.2 | 3.1 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.2 | 4.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.0 | GO:0060136 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 18.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 2.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 4.9 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 16.2 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 2.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 4.7 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 2.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 3.8 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 45.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 9.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 3.7 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 0.3 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 1.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 2.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 8.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.7 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.9 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 2.3 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 1.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 3.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 7.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.7 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.4 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 6.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 2.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 2.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 2.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 2.4 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 2.3 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 2.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) |
| 0.0 | 0.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 2.0 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 3.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 35.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 6.6 | 19.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 5.7 | 113.4 | GO:0042555 | MCM complex(GO:0042555) |
| 5.4 | 53.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 4.9 | 19.8 | GO:0000811 | GINS complex(GO:0000811) |
| 4.7 | 18.7 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 2.6 | 18.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 2.1 | 17.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.8 | 1.8 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 1.8 | 18.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.7 | 20.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.6 | 9.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.6 | 15.6 | GO:0000796 | condensin complex(GO:0000796) |
| 1.5 | 4.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.5 | 15.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.5 | 8.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.3 | 49.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 1.1 | 6.9 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.1 | 5.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 1.1 | 12.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.1 | 14.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.0 | 3.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 1.0 | 3.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.0 | 2.9 | GO:0005663 | DNA replication factor C complex(GO:0005663) PCNA-p21 complex(GO:0070557) |
| 0.9 | 3.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.8 | 5.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.8 | 31.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.8 | 13.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.6 | 9.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.6 | 3.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.5 | 32.8 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.5 | 11.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 8.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 3.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 6.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 1.7 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.4 | 1.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.4 | 1.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 1.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 7.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 4.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 3.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 8.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 0.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.2 | 2.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 2.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 2.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 8.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 30.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 2.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 7.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 4.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 24.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 12.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 9.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 47.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.8 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 10.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 2.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 2.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 23.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 3.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 2.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 3.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 4.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.4 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 4.6 | 13.7 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 3.9 | 23.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 2.9 | 66.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 2.8 | 11.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 2.2 | 17.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 2.1 | 17.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.6 | 6.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 1.5 | 30.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.5 | 9.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.4 | 35.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 1.3 | 19.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 1.3 | 20.9 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 1.3 | 5.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.2 | 11.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.1 | 5.7 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 1.1 | 7.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.1 | 12.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.0 | 3.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 1.0 | 8.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.0 | 37.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 1.0 | 36.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.9 | 5.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.8 | 7.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.8 | 4.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.7 | 2.9 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.7 | 3.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.7 | 2.0 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.5 | 3.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.5 | 1.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.5 | 1.6 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.4 | 2.5 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.4 | 3.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.4 | 17.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 1.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.3 | 2.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 28.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 23.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.3 | 1.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 44.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 10.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 2.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 1.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 1.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 2.1 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.7 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 1.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.4 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 47.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.0 | GO:0016174 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 3.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 18.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 4.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 16.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 3.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 14.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 20.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 7.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 4.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 3.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 2.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.8 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 3.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 16.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 3.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 4.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 12.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 11.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 6.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 18.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 4.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 2.0 | GO:0015399 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 3.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 3.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0070717 | poly-purine tract binding(GO:0070717) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 48.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.5 | 39.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.9 | 70.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.8 | 19.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.8 | 36.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 45.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 29.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 29.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 9.5 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 20.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 15.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 2.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 6.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 9.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 9.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 11.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 6.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 3.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 133.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 3.5 | 112.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 2.4 | 21.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.8 | 12.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 1.2 | 9.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.0 | 19.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.0 | 23.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.9 | 29.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.9 | 20.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.9 | 20.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 30.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.5 | 4.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 32.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.4 | 18.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 13.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 22.1 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.3 | 13.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 11.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 12.7 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.3 | 4.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 9.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 3.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 6.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 4.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 7.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 9.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 7.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.6 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 6.9 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 4.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 6.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 19.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 2.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |