Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
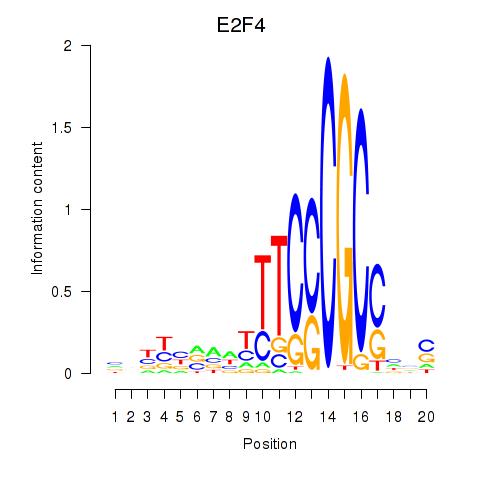
Results for E2F4
Z-value: 3.51
Transcription factors associated with E2F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F4
|
ENSG00000205250.4 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F4 | hg19_v2_chr16_+_67226019_67226127 | 0.62 | 3.5e-03 | Click! |
Activity profile of E2F4 motif
Sorted Z-values of E2F4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 2.9 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 2.5 | 7.5 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 1.8 | 5.4 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.6 | 4.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 1.6 | 6.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 1.3 | 9.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.1 | 3.4 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 1.1 | 9.0 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.1 | 4.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.1 | 2.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 1.0 | 4.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 1.0 | 3.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.0 | 5.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 1.0 | 2.9 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.0 | 2.9 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.9 | 2.6 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.8 | 9.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.8 | 5.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.8 | 2.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.7 | 5.2 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 2.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.7 | 2.2 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.7 | 3.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.6 | 2.6 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.6 | 4.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.6 | 1.7 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.5 | 1.6 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.5 | 2.6 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.5 | 1.5 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.5 | 1.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.5 | 7.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 1.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.4 | 1.8 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.4 | 2.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.4 | 1.3 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.4 | 5.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 1.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.4 | 2.6 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.4 | 2.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.4 | 2.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.4 | 2.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.3 | 3.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.7 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.3 | 2.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.0 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.3 | 1.0 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 1.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.3 | 0.9 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.3 | 0.6 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.3 | 3.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 0.9 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.3 | 1.6 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.3 | 1.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.3 | 2.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 1.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 2.0 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.2 | 2.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 2.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 1.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 9.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 2.6 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 3.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 6.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 2.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 0.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 1.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.2 | 4.7 | GO:0009650 | UV protection(GO:0009650) |
| 0.2 | 1.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 2.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 1.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 1.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 0.7 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 1.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 3.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 0.8 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.2 | 1.7 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 2.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 1.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.6 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 2.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 4.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 2.3 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 1.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 7.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 2.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 3.1 | GO:0000732 | strand displacement(GO:0000732) |
| 0.1 | 4.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 6.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 5.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 1.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 2.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 1.3 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.3 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.7 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 1.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 2.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 1.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.1 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.9 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.4 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 5.1 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 1.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 2.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.9 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 2.8 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.1 | 1.4 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.5 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 2.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 1.2 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.6 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 1.8 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 6.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 1.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.5 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 1.1 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.5 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 2.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 1.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.8 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 2.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 3.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.4 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.9 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 1.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 2.8 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.8 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 1.6 | 4.7 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.9 | 2.8 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.9 | 8.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.7 | 2.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 8.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 2.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.6 | 2.9 | GO:0071458 | polycystin complex(GO:0002133) integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.6 | 4.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.6 | 1.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.6 | 1.7 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.5 | 2.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 9.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 1.5 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.4 | 9.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.4 | 8.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.4 | 2.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 1.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.4 | 3.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 4.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 2.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.4 | 1.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 2.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 5.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 2.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 2.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 4.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 3.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 1.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 5.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 1.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 2.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.9 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 1.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.9 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 2.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 6.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 5.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 0.6 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 1.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 2.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 3.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 2.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 9.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 5.7 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 1.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 3.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 6.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 4.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 4.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 4.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 2.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 1.3 | 4.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 1.1 | 3.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.1 | 5.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 1.0 | 3.9 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.0 | 9.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.9 | 5.2 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.9 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.8 | 2.3 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.8 | 5.4 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.7 | 2.9 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.7 | 2.2 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.7 | 6.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.7 | 4.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.6 | 2.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.6 | 1.9 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.6 | 2.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.6 | 2.2 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.6 | 3.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.5 | 1.6 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.5 | 2.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.5 | 4.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 1.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.4 | 1.3 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.4 | 1.7 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.4 | 1.3 | GO:0032143 | single guanine insertion binding(GO:0032142) single thymine insertion binding(GO:0032143) |
| 0.4 | 3.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 1.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 5.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 2.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.3 | 1.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 2.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 1.8 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.3 | 0.8 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.3 | 6.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 7.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 2.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.9 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.2 | 4.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 1.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.2 | 0.8 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 1.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 10.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 1.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 3.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 1.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 0.7 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.2 | 2.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 1.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 14.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 3.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.5 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 2.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 2.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.6 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 2.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 3.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 3.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.4 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 1.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 2.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 2.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.2 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 1.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 3.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 7.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 5.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 4.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 3.0 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 1.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.0 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.8 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 1.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 5.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 9.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.0 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 36.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 14.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 7.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 5.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 2.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 6.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 2.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.7 | 13.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.7 | 8.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.6 | 11.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 19.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 3.8 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.3 | 1.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.3 | 7.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 7.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 8.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 2.9 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 6.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 13.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 8.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 6.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 5.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 13.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 4.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 5.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 3.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 4.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 6.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |