Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for E2F8
Z-value: 2.33
Transcription factors associated with E2F8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F8
|
ENSG00000129173.8 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F8 | hg19_v2_chr11_-_19262486_19262512 | 0.78 | 6.0e-05 | Click! |
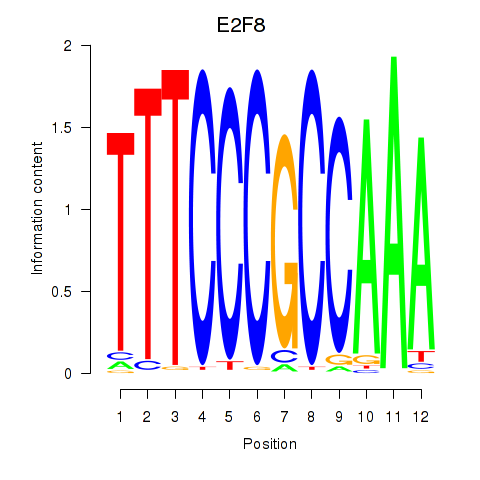
Activity profile of E2F8 motif
Sorted Z-values of E2F8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.9 | 11.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.1 | 3.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.0 | 3.0 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.8 | 9.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.7 | 9.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.7 | 2.1 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.7 | 4.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.7 | 0.7 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.7 | 2.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.6 | 1.9 | GO:0071812 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.6 | 4.4 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.5 | 2.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.5 | 8.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 2.9 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.5 | 1.4 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.5 | 1.4 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.4 | 2.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.4 | 12.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 1.8 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.4 | 1.1 | GO:0035573 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.3 | 1.4 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.3 | 1.3 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 4.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 | 2.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 2.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 1.9 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 4.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 0.8 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 0.8 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.3 | 1.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.3 | 2.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 3.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 1.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 11.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 0.9 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 2.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 1.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) protein K6-linked ubiquitination(GO:0085020) |
| 0.2 | 3.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 6.1 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.2 | 0.8 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 1.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.2 | 3.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.2 | 3.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.2 | 1.0 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.2 | 0.5 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.2 | 1.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 3.5 | GO:0000732 | strand displacement(GO:0000732) |
| 0.1 | 0.6 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 1.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.6 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.7 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.8 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 1.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.5 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 2.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 5.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 1.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.9 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) mRNA methylation(GO:0080009) |
| 0.1 | 0.8 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 2.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.3 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 2.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 2.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 0.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.5 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 2.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.6 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.8 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 3.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 1.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.9 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 2.3 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 3.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.3 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 1.6 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 2.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 5.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.7 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.6 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.8 | GO:0009223 | pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 2.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.1 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.3 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 1.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.2 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.8 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0031523 | Myb complex(GO:0031523) |
| 1.1 | 4.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.1 | 3.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.9 | 18.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.9 | 5.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.7 | 5.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.7 | 2.6 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.5 | 1.9 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.5 | 2.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 8.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 5.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 12.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 1.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 2.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 1.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.3 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 1.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 3.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 5.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 9.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 2.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 2.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 2.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 4.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 1.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 2.0 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 2.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 2.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 3.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 5.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 4.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 4.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0030863 | COP9 signalosome(GO:0008180) cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 1.1 | 4.6 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 1.0 | 4.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.9 | 4.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.8 | 6.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.8 | 17.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.7 | 3.7 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.5 | 3.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 1.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.4 | 1.2 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.3 | 1.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.3 | 1.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 2.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 0.8 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.3 | 2.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.3 | 0.8 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.3 | 1.5 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.2 | 1.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 1.8 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.7 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 3.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 0.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 1.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 2.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.6 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.2 | 8.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 2.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.8 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 2.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 3.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 1.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 2.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.4 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 10.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.2 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 4.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 2.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 1.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 1.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 7.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 2.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 3.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.8 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 4.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 2.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 2.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 7.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 12.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 4.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 21.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 8.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 4.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 4.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 3.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 6.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 5.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 4.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 3.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 3.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |