Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
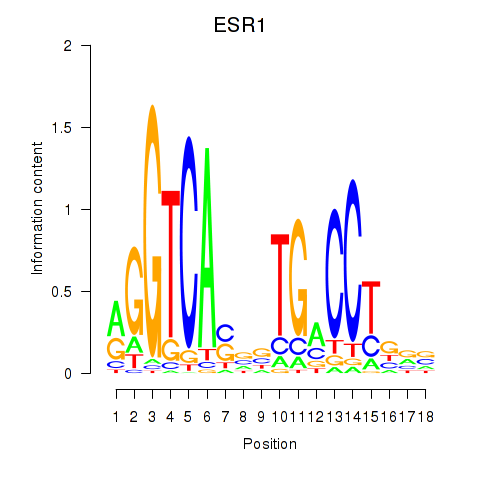
Results for ESR1
Z-value: 2.24
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.17 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg19_v2_chr6_+_152128371_152128454 | -0.84 | 3.5e-06 | Click! |
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0100012 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 1.4 | 7.0 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.3 | 4.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 1.3 | 7.7 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 1.1 | 5.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.1 | 3.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.0 | 5.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.9 | 3.8 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.8 | 2.3 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.7 | 1.3 | GO:0019229 | regulation of vasoconstriction(GO:0019229) positive regulation of vasoconstriction(GO:0045907) |
| 0.7 | 2.0 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.6 | 2.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.6 | 1.8 | GO:0060929 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.6 | 1.2 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.6 | 1.7 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.6 | 2.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.5 | 2.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.5 | 1.9 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.5 | 1.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.5 | 1.4 | GO:1903216 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.4 | 1.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 1.8 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.4 | 2.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.4 | 1.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.4 | 0.4 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.4 | 2.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.4 | 1.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 1.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.4 | 3.0 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.4 | 10.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.4 | 1.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.4 | 4.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 2.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 1.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 1.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 1.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.3 | 1.5 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.3 | 4.8 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 1.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.3 | 2.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.3 | 1.8 | GO:0046618 | drug export(GO:0046618) |
| 0.3 | 2.4 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.3 | 0.8 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.3 | 2.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 6.2 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.3 | 4.6 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.3 | 1.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.3 | 2.6 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.3 | 0.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 0.8 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 1.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.3 | 3.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 3.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 2.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 1.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 3.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 4.9 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.6 | GO:0036102 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.2 | 2.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.2 | 3.5 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 0.4 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.2 | 5.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 2.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 0.9 | GO:2000230 | response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 2.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 3.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.4 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.2 | 0.9 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 5.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.2 | 1.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 0.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 4.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 0.6 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.2 | 0.4 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.2 | 0.9 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.2 | 1.3 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.2 | 0.9 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.2 | 1.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 1.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.2 | 0.8 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.2 | 1.7 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.2 | 1.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 1.5 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.2 | 0.8 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.2 | 0.9 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 0.5 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.2 | 0.6 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 1.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.4 | GO:1900365 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) positive regulation of mRNA polyadenylation(GO:1900365) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 3.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 1.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.3 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.8 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.4 | GO:0035566 | vestibulocochlear nerve formation(GO:0021650) regulation of metanephros size(GO:0035566) |
| 0.1 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.5 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.1 | 0.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.6 | GO:1990569 | GDP-fucose transport(GO:0015783) UDP-N-acetylglucosamine transport(GO:0015788) purine nucleotide-sugar transport(GO:0036079) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.6 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.1 | 0.5 | GO:0043366 | beta selection(GO:0043366) |
| 0.1 | 0.5 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.8 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.7 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 2.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.4 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.1 | 2.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 4.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.8 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 2.6 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.5 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 | 0.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 1.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.4 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 1.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.5 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.4 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 3.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.1 | 0.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 1.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 1.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.2 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.1 | 7.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.4 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 2.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.5 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.5 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.1 | 1.5 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 0.3 | GO:0090034 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 0.6 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 2.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 3.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.6 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.5 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.9 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.3 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 1.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.8 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.5 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.4 | GO:1990253 | cellular response to leucine(GO:0071233) cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.8 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.7 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.8 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.1 | GO:0019860 | uracil metabolic process(GO:0019860) |
| 0.1 | 0.3 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.8 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.3 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.4 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 1.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 1.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.5 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 1.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 2.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 2.4 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.3 | GO:0048672 | abscission(GO:0009838) regulation of collateral sprouting(GO:0048670) positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.7 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.6 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.8 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 7.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.5 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.5 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.8 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 2.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0006863 | purine nucleobase transport(GO:0006863) |
| 0.0 | 1.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 2.1 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.5 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.0 | 0.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 2.7 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.8 | GO:0040008 | regulation of growth(GO:0040008) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.5 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.1 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 1.3 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.9 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 1.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 1.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:2000977 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) primitive erythrocyte differentiation(GO:0060319) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.0 | 1.5 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 2.0 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 1.0 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 1.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.6 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.1 | GO:0097205 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.7 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 0.5 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.0 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.4 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.5 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.6 | 3.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.6 | 2.2 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.6 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 7.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.4 | 1.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.4 | 4.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 0.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.3 | 1.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 3.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 3.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 6.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 1.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 8.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 1.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 2.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 2.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 1.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.6 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 4.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.8 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 4.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 1.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 3.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.7 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 3.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 9.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 1.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.8 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 3.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 2.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 2.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 4.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 8.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.5 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 3.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 3.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.9 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 1.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 2.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 4.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.6 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 2.9 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 25.4 | GO:0044429 | mitochondrial part(GO:0044429) |
| 0.0 | 3.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.2 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 2.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.5 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.8 | 3.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.7 | 2.0 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.6 | 3.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.6 | 1.7 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.6 | 7.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 5.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.5 | 1.6 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.5 | 2.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.5 | 1.6 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 3.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 1.4 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.5 | 1.4 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.4 | 1.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 1.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.4 | 3.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.4 | 8.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 4.8 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 1.5 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.4 | 1.8 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.4 | 6.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.3 | 2.4 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 4.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 2.6 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 0.9 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.3 | 1.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 1.8 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.3 | 6.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 2.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 4.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.3 | 0.8 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 0.8 | GO:0008124 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.3 | 1.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 2.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 0.7 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 3.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 11.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.0 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.2 | 0.8 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 1.8 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.7 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.2 | 5.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.8 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.2 | 3.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.5 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.2 | 1.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 0.8 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.2 | 0.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 0.6 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 1.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 2.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.5 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 1.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 4.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.6 | GO:0005462 | GDP-fucose transmembrane transporter activity(GO:0005457) UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 3.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.4 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.8 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.6 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 3.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 5.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.5 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.4 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.4 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 2.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 2.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.0 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 3.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 8.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 1.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.7 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 2.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 2.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 1.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 4.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 4.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 2.5 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 3.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0004104 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.4 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 1.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 5.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 5.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 3.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 3.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 3.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 4.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 6.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 4.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 4.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 12.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 3.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 6.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 8.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 2.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 5.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 3.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 3.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 2.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 2.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 2.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.9 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 3.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 4.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 3.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.7 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |