Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
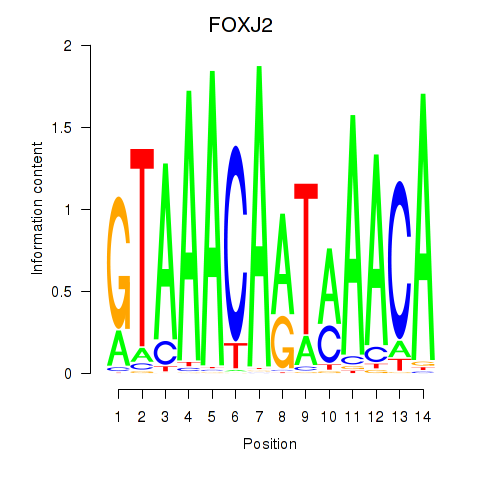
Results for FOXJ2
Z-value: 1.97
Transcription factors associated with FOXJ2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXJ2
|
ENSG00000065970.4 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ2 | hg19_v2_chr12_+_8185288_8185339 | 0.73 | 2.5e-04 | Click! |
Activity profile of FOXJ2 motif
Sorted Z-values of FOXJ2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXJ2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.4 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 1.3 | 5.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 1.2 | 3.6 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 1.1 | 4.2 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.6 | 3.7 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.5 | 1.6 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.5 | 3.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.4 | 3.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 2.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.4 | 3.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.4 | 1.5 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.4 | 1.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.4 | 3.9 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 2.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 | 5.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 4.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 1.6 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 2.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 1.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 1.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 1.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 1.3 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.2 | 3.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 9.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 2.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 1.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.3 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 3.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.7 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.9 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.0 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 3.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 2.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 6.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 1.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.7 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 1.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.8 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.9 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 1.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.7 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 3.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 1.3 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 3.9 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.6 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 1.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 1.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.8 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.3 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.5 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 3.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 4.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 8.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 2.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 0.9 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.3 | 2.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 0.8 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 1.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 3.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 2.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 8.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 0.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 3.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.8 | GO:0098862 | brush border(GO:0005903) cluster of actin-based cell projections(GO:0098862) |
| 0.1 | 2.5 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 3.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.9 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 0.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.8 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 3.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.9 | 3.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.4 | 1.3 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.4 | 3.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.4 | 3.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.4 | 1.5 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.3 | 0.8 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.2 | 0.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 7.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.7 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 3.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 2.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 3.2 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.1 | 1.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 3.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.1 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 1.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 2.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 10.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.0 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.5 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 3.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 5.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 3.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 10.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 4.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 3.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 8.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 11.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 3.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 3.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 4.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |