Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
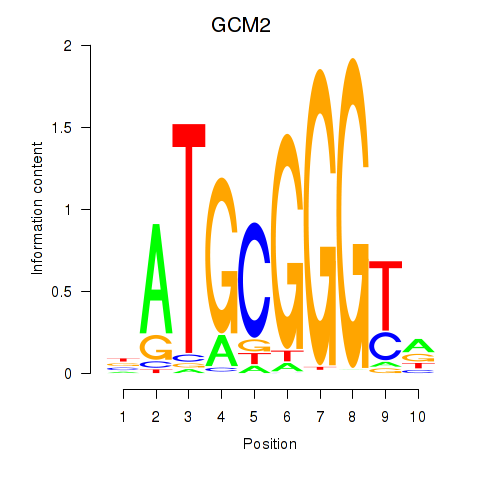
Results for GCM2
Z-value: 2.64
Transcription factors associated with GCM2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCM2
|
ENSG00000124827.6 | glial cells missing transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GCM2 | hg19_v2_chr6_-_10882174_10882274 | 0.12 | 6.0e-01 | Click! |
Activity profile of GCM2 motif
Sorted Z-values of GCM2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GCM2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.4 | 5.5 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.0 | 4.2 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.8 | 4.7 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.8 | 2.3 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.7 | 2.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.7 | 2.6 | GO:0023016 | osmosensory signaling pathway(GO:0007231) signal transduction by trans-phosphorylation(GO:0023016) kidney smooth muscle tissue development(GO:0072194) pattern specification involved in metanephros development(GO:0072268) |
| 0.6 | 3.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.6 | 1.9 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.6 | 3.9 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.6 | 2.8 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.6 | 1.7 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.6 | 2.2 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.5 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.5 | 2.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.5 | 5.2 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.5 | 1.9 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.4 | 4.9 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.4 | 3.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.4 | 1.6 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.4 | 4.6 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.4 | 1.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.3 | 2.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.3 | 1.0 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.3 | 3.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 5.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 0.9 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 3.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.3 | 7.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.3 | 0.8 | GO:1903414 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.3 | 0.8 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.3 | 1.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.2 | 1.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.7 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 3.4 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.2 | 5.8 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 1.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.2 | 0.6 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 0.4 | GO:0071661 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.2 | 0.7 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.2 | 0.5 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.2 | 1.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.2 | 5.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 2.7 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.2 | 3.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.4 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.1 | 0.9 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 3.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 3.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.4 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 1.8 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.4 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 2.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.0 | GO:0060501 | negative regulation of keratinocyte differentiation(GO:0045617) positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.7 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 1.0 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.6 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 1.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 1.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 1.0 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.5 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 1.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 3.3 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.1 | 0.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 1.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 1.6 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.1 | 1.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.5 | GO:0071105 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 1.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.9 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 2.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 1.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 2.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.0 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.6 | GO:0006538 | glutamate biosynthetic process(GO:0006537) glutamate catabolic process(GO:0006538) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 2.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.5 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.5 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 3.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 2.4 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.0 | 1.8 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.3 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 3.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.8 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 1.9 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 1.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 4.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 2.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.0 | 1.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.4 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.9 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 1.6 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 2.7 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 1.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.2 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 11.1 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.5 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 2.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.8 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 2.1 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.7 | 5.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 2.7 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.4 | 3.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 3.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 1.0 | GO:0097124 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 2.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 1.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 3.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 5.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 3.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.1 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 3.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 5.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 11.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 2.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 4.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 2.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 1.7 | 5.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.2 | 7.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.9 | 4.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.8 | 3.8 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.6 | 3.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 2.3 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.4 | 3.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.4 | 15.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 4.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 3.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 3.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 5.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.9 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.3 | 2.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.3 | 1.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.3 | 3.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 2.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 1.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.3 | 1.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 4.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 0.8 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.3 | 2.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 1.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 1.0 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 0.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.7 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 5.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 5.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 0.5 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.2 | 3.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 3.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.6 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 3.2 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.6 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.1 | 0.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.9 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 4.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 2.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 1.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 4.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 2.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 1.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.5 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.8 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 2.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.5 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 3.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 6.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 6.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.1 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 2.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 1.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 20.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 3.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 4.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 4.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 10.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 4.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 15.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 3.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 3.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 7.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 3.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 5.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 2.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 5.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 2.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.2 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |