Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
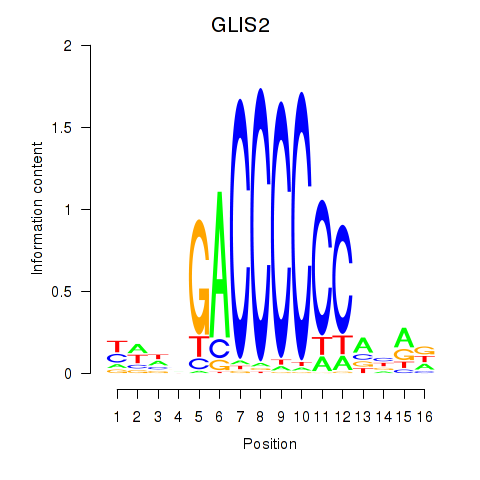
Results for GLIS2
Z-value: 3.31
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.4 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS2 | hg19_v2_chr16_+_4382225_4382225 | 0.86 | 1.2e-06 | Click! |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 28.9 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 4.3 | 12.9 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 4.1 | 12.4 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 2.3 | 9.1 | GO:0060032 | notochord regression(GO:0060032) |
| 2.1 | 6.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 1.9 | 5.8 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 1.9 | 5.6 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 1.7 | 5.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 1.5 | 9.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 1.3 | 11.7 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 1.2 | 4.7 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.8 | 3.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.8 | 2.3 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.7 | 2.1 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.7 | 2.0 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.7 | 2.0 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.6 | 2.6 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.6 | 2.4 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.6 | 10.0 | GO:0030238 | male sex determination(GO:0030238) |
| 0.6 | 1.7 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.6 | 1.7 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.5 | 1.6 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 | 2.4 | GO:0003277 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.5 | 1.8 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.4 | 1.3 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 | 3.0 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 1.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.4 | 1.2 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.4 | 2.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.4 | 8.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.4 | 3.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.3 | 1.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.3 | 1.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.3 | 1.7 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.3 | 11.1 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.3 | 0.3 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.3 | 2.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 1.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.3 | 2.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 6.1 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.2 | 0.9 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 0.9 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.2 | 6.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 2.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 16.9 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 0.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 0.8 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 3.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 4.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.7 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.2 | 3.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 2.2 | GO:0043951 | negative regulation of cell adhesion mediated by integrin(GO:0033629) negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 3.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.5 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.2 | 2.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 2.8 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.2 | 1.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.2 | 4.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.2 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.2 | 1.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 2.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 5.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 3.4 | GO:0050961 | cell-cell adhesion mediated by cadherin(GO:0044331) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 1.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.6 | GO:0060316 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 3.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.1 | 2.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.7 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 8.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.3 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.1 | 1.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 2.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 2.2 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.1 | 0.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 2.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.3 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 1.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.3 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.4 | GO:2000866 | negative regulation of macrophage chemotaxis(GO:0010760) positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 2.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.5 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 1.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 0.3 | GO:0034699 | response to luteinizing hormone(GO:0034699) |
| 0.1 | 0.4 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 1.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 12.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.2 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 2.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 14.8 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.1 | 0.4 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 2.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.8 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.1 | 0.6 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.1 | 1.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.9 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 6.5 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.3 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 2.3 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 1.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 1.7 | GO:0090383 | ATP hydrolysis coupled proton transport(GO:0015991) phagosome acidification(GO:0090383) |
| 0.0 | 8.8 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 1.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 2.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 1.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 2.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.9 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 2.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 1.0 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 2.0 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 2.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 5.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.0 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.9 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 4.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:1901800 | positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 1.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 2.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.5 | GO:1901343 | negative regulation of vasculature development(GO:1901343) |
| 0.0 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.8 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.9 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.6 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.4 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.9 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) |
| 0.0 | 1.9 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 1.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 1.0 | GO:0070527 | platelet aggregation(GO:0070527) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 1.2 | 2.4 | GO:0019034 | viral replication complex(GO:0019034) |
| 1.0 | 26.6 | GO:0042627 | chylomicron(GO:0042627) |
| 1.0 | 5.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.9 | 2.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.7 | 9.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.6 | 2.6 | GO:1990742 | microvesicle(GO:1990742) |
| 0.6 | 3.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.6 | 12.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 3.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 6.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 2.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 2.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.4 | 3.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 3.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 2.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 8.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 3.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 1.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.3 | 1.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 4.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 3.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 3.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 4.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 0.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 11.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 2.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 0.7 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 2.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 0.9 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.2 | 7.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 16.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 0.7 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 1.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 3.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 2.9 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.8 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 23.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 8.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 2.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.2 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 2.1 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 1.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 3.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 2.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 6.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 6.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.7 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 15.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 2.7 | 26.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.9 | 4.6 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.8 | 2.3 | GO:0047726 | iron-cytochrome-c reductase activity(GO:0047726) |
| 0.7 | 2.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.7 | 18.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 2.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.7 | 2.0 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.6 | 8.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 1.7 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.5 | 1.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.4 | 2.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.4 | 1.6 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.4 | 12.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 3.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 8.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 12.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 5.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.3 | 2.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 2.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 1.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.3 | 1.6 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.3 | 1.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.3 | 3.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 3.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 10.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.2 | 2.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 6.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 2.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 2.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 0.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 2.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.6 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 1.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 4.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 3.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.8 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 1.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.3 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.1 | 2.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 5.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 3.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.8 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 1.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.1 | GO:0052658 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.4 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.4 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 11.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 1.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.3 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 1.2 | GO:0046961 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 5.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 8.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 7.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 2.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 1.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 15.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 14.3 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 3.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin-protein transferase regulator activity(GO:0055106) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 6.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 2.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 5.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 2.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 5.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 8.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 18.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 7.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 5.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 3.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 12.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 6.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 6.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 5.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.7 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 8.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 4.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 9.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 10.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 11.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 7.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 3.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 4.4 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 2.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 3.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |