Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
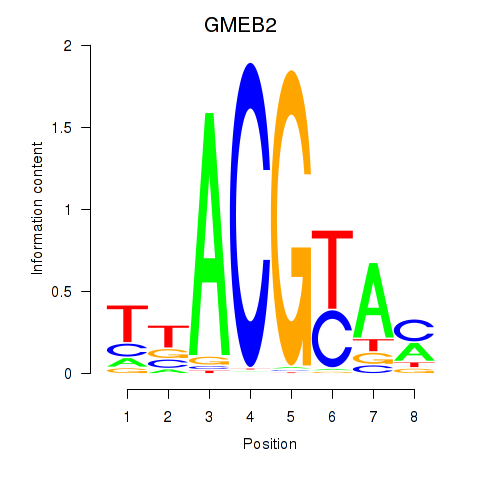
Results for GMEB2
Z-value: 3.33
Transcription factors associated with GMEB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GMEB2
|
ENSG00000101216.6 | glucocorticoid modulatory element binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | hg19_v2_chr20_-_62258394_62258464 | 0.18 | 4.4e-01 | Click! |
Activity profile of GMEB2 motif
Sorted Z-values of GMEB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GMEB2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.5 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 1.8 | 5.3 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 1.5 | 4.5 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.9 | 2.8 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.9 | 3.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.8 | 2.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.8 | 3.0 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.7 | 4.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.6 | 2.5 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.6 | 5.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.6 | 2.5 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.6 | 3.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.6 | 11.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 2.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.6 | 1.8 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.6 | 1.7 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 | 2.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.5 | 2.7 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.5 | 1.6 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.5 | 6.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.5 | 3.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 2.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.5 | 2.0 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.5 | 4.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 4.7 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.4 | 1.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.4 | 2.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 1.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.4 | 2.8 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.4 | 3.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.4 | 2.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.4 | 1.5 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.4 | 3.0 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.4 | 2.3 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.4 | 2.8 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.4 | 2.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.4 | 1.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 1.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 1.7 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.3 | 1.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.3 | 1.0 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.3 | 3.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.3 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.3 | 1.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 1.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.3 | 1.6 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.3 | 4.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 0.9 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.3 | 1.8 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.3 | 1.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 2.8 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.3 | 0.8 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 | 1.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.3 | 5.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 4.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 0.8 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.3 | 1.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 0.8 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.3 | 0.8 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.3 | 1.3 | GO:1904751 | positive regulation of telomeric DNA binding(GO:1904744) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.3 | 0.8 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.3 | 1.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 2.0 | GO:0042256 | mature ribosome assembly(GO:0042256) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.2 | 1.5 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 4.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 2.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.2 | GO:0035698 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of T cell extravasation(GO:2000407) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.2 | 5.4 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.2 | 1.9 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 0.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 1.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.2 | 0.9 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 0.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.8 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 | 2.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 1.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.2 | 1.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.2 | 1.3 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 2.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 0.9 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 1.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 1.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 3.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 1.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 2.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 3.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 2.0 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 0.8 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 1.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 1.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 5.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 2.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 3.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 1.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 0.6 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 0.7 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.2 | 2.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.2 | 0.9 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 | 0.5 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 2.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.7 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.2 | 1.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.2 | 0.5 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.2 | 2.0 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 0.5 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.2 | 0.7 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 | 0.5 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 0.8 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.2 | 0.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 | 1.6 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 2.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 2.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.2 | 3.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 1.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 0.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.3 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.7 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 7.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 1.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 4.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 3.8 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.1 | 0.4 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 0.7 | GO:2000255 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 1.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 2.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 3.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 1.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 5.8 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.3 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 2.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.8 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.4 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 1.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.9 | GO:0070970 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) interleukin-2 secretion(GO:0070970) |
| 0.1 | 1.7 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.9 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.1 | 1.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.5 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.5 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 2.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.1 | GO:0046102 | adenosine catabolic process(GO:0006154) hypoxanthine salvage(GO:0043103) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 1.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 2.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.9 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.1 | 0.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.9 | GO:0035898 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.1 | 0.8 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.1 | 0.3 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 0.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.6 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.4 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 1.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 1.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 1.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.8 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.1 | 0.3 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.1 | 1.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 2.4 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.1 | 3.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.3 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 1.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 1.2 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
| 0.1 | 0.3 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.1 | 1.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 2.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 1.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.1 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.1 | 2.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 1.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 4.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.6 | GO:0071901 | negative regulation of protein serine/threonine kinase activity(GO:0071901) |
| 0.1 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 2.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.4 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.8 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.8 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 1.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 | 1.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 1.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.1 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.1 | 0.2 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.2 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.7 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 3.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.4 | GO:0060846 | blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 1.7 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 1.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 2.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 1.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 1.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 2.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 3.7 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 1.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 1.8 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 1.6 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 3.6 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 2.4 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 1.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.8 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) regulation of GTP binding(GO:1904424) |
| 0.0 | 3.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.5 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 1.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.3 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 3.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.4 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.9 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.9 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.9 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 4.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 5.3 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 3.8 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.5 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 2.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.8 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.4 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.5 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.5 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.6 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 2.7 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 2.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.9 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.7 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.6 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.0 | 0.9 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:2001171 | regulation of ATP biosynthetic process(GO:2001169) positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 2.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.6 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.5 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:1902099 | regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.0 | 1.1 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.9 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.8 | 11.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.7 | 2.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.7 | 4.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.6 | 3.8 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.5 | 4.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.5 | 3.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.5 | 6.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.5 | 4.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.5 | 1.4 | GO:0097134 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.5 | 1.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 2.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 1.3 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.4 | 7.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 1.5 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.4 | 1.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 1.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 1.0 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.3 | 2.2 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 2.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 4.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 1.5 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.3 | 0.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.3 | 1.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.3 | 1.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 1.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 1.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 8.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 5.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 0.9 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 0.6 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 2.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 3.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.2 | 3.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 1.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 1.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 3.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.8 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 4.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 2.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.7 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.1 | 1.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 3.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.9 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 1.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 2.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.3 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 1.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.4 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.1 | 2.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0019012 | virion(GO:0019012) virion part(GO:0044423) |
| 0.1 | 1.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 2.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 2.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.8 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.1 | 0.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 10.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 2.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 2.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 0.5 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 1.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 6.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 3.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.2 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 0.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 5.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 7.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 2.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 4.8 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.0 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.7 | GO:0030684 | preribosome(GO:0030684) |
| 0.0 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.8 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 3.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 3.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 9.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 8.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 2.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.7 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 1.8 | 5.3 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.8 | 0.8 | GO:0017076 | purine nucleotide binding(GO:0017076) |
| 0.7 | 3.7 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.7 | 2.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.7 | 2.8 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.6 | 2.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.6 | 3.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 2.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.6 | 1.8 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.5 | 2.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.5 | 1.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.4 | 3.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 3.6 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.4 | 1.7 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.4 | 1.7 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.4 | 2.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.4 | 5.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 2.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.4 | 2.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.4 | 1.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.4 | 1.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.4 | 4.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 1.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 1.0 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.3 | 1.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 1.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.3 | 3.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 15.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 0.9 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.3 | 1.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.3 | 5.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 1.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.3 | 1.2 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.3 | 2.4 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 1.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 1.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 0.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 0.8 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.3 | 0.8 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.3 | 1.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 0.8 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 2.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 5.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 0.9 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.2 | 4.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 6.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 2.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.2 | 2.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.2 | 1.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 1.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 1.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 1.8 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.2 | 1.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 2.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 3.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.6 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 1.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.6 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.2 | 2.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 2.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 6.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 0.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 1.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 1.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 0.8 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.2 | 1.0 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.2 | 4.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 0.8 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.2 | 2.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.2 | GO:0015186 | L-cystine transmembrane transporter activity(GO:0015184) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 5.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.9 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 1.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 4.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 4.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 3.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 4.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 3.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.5 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 3.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 5.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 4.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.6 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.6 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 2.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 5.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 2.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 4.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.3 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 3.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 2.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 7.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 3.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.0 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 2.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 5.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.1 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 2.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.2 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 2.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.6 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 3.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 2.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 2.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 1.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 2.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0016301 | kinase activity(GO:0016301) |
| 0.0 | 5.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 6.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 8.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 15.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 8.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 7.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 5.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 5.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 3.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 3.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 3.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 4.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 14.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 11.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 3.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 8.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 4.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 0.5 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 6.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 7.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 9.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 4.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 5.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 3.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 3.0 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.1 | 2.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.7 | REACTOME METABOLISM OF RNA | Genes involved in Metabolism of RNA |
| 0.1 | 0.8 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.1 | 4.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 4.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 8.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 3.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 3.7 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 3.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 4.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 6.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |