Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
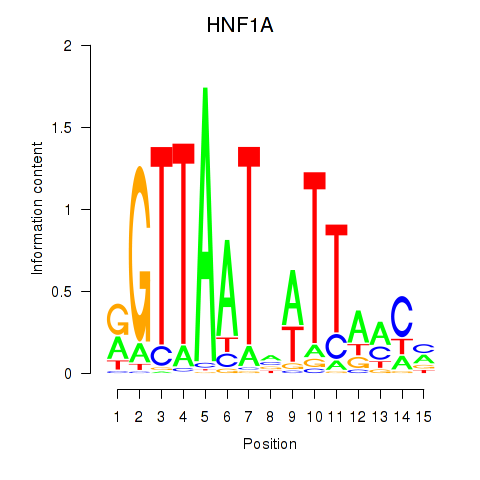
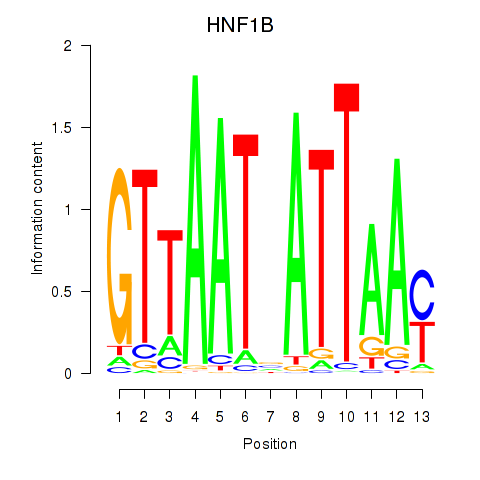
Results for HNF1A_HNF1B
Z-value: 8.91
Transcription factors associated with HNF1A_HNF1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF1A
|
ENSG00000135100.13 | HNF1 homeobox A |
|
HNF1B
|
ENSG00000108753.8 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF1A | hg19_v2_chr12_+_121416340_121416371 | 0.94 | 5.9e-10 | Click! |
| HNF1B | hg19_v2_chr17_-_36105009_36105060 | 0.82 | 1.1e-05 | Click! |
Activity profile of HNF1A_HNF1B motif
Sorted Z-values of HNF1A_HNF1B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF1A_HNF1B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 41.9 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 13.3 | 79.7 | GO:1903412 | response to bile acid(GO:1903412) |
| 9.2 | 27.6 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 8.1 | 105.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 7.9 | 110.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 6.8 | 20.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 6.1 | 36.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 5.2 | 36.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 4.8 | 48.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 4.6 | 13.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 4.3 | 51.4 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 4.2 | 8.5 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 4.0 | 19.8 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 3.4 | 13.7 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 3.4 | 10.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 3.1 | 31.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 3.1 | 30.7 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 2.8 | 16.7 | GO:0051958 | methotrexate transport(GO:0051958) |
| 2.0 | 5.9 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 2.0 | 7.9 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 1.9 | 11.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.8 | 9.0 | GO:2000005 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 1.8 | 5.3 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 1.7 | 7.0 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 1.6 | 4.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.5 | 4.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 1.4 | 28.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 1.4 | 47.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 1.4 | 14.9 | GO:0015747 | urate transport(GO:0015747) |
| 1.3 | 3.9 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 1.2 | 9.6 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 1.2 | 7.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 1.0 | 10.4 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.9 | 7.4 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.9 | 11.3 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.8 | 2.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.8 | 2.3 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.8 | 8.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.7 | 2.2 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.7 | 4.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.6 | 20.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.6 | 15.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.6 | 5.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.5 | 2.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.5 | 3.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 2.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 16.8 | GO:0060065 | uterus development(GO:0060065) |
| 0.4 | 1.6 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.4 | 3.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.3 | 32.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.3 | 3.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 17.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.3 | 3.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 8.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 5.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 2.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 36.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.2 | 2.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 3.8 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 2.6 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 2.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 2.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 8.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 1.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.2 | 8.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.2 | 18.9 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.1 | 0.6 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.7 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 4.7 | GO:0010827 | regulation of glucose transport(GO:0010827) regulation of glucose import(GO:0046324) |
| 0.1 | 0.7 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.4 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 1.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 4.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 1.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 9.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 14.6 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 2.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 4.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 5.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 8.4 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 18.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 1.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 2.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 9.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 6.6 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.8 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 13.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 7.1 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 2.3 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 2.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 2.4 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 5.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 4.9 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 132.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 4.6 | 36.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 3.3 | 39.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 2.6 | 46.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.8 | 5.3 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 1.7 | 11.7 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 1.3 | 3.8 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.3 | 31.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 1.1 | 28.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.9 | 15.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 4.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 4.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.5 | 8.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.4 | 2.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 4.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.3 | 3.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 3.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 13.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 5.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 13.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 30.7 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.2 | 18.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 18.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 8.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 4.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 4.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 79.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 2.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 48.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 7.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 11.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 105.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 40.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 9.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 58.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 29.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 11.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 3.2 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 7.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 3.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 21.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 16.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 5.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 27.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 7.5 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 26.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 47.4 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 46.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 9.2 | 27.6 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 9.1 | 36.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 8.8 | 105.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 6.6 | 79.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 6.6 | 19.8 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 4.6 | 13.9 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 4.1 | 20.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.4 | 13.7 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 3.4 | 47.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 3.2 | 35.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 2.8 | 16.7 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 2.2 | 41.1 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 1.7 | 11.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.6 | 4.8 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 1.4 | 17.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.3 | 37.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.3 | 5.3 | GO:0070326 | very-low-density lipoprotein particle binding(GO:0034189) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 1.3 | 28.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.3 | 14.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.2 | 8.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.1 | 7.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 1.0 | 7.3 | GO:0016160 | amylase activity(GO:0016160) |
| 1.0 | 31.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.9 | 2.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.9 | 4.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.9 | 11.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.9 | 2.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.9 | 2.6 | GO:0008124 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.8 | 36.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.8 | 9.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.8 | 46.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.7 | 18.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.7 | 5.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.7 | 7.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.6 | 9.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.6 | 3.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.6 | 2.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.6 | 2.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.6 | 3.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.5 | 10.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 4.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 4.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.4 | 70.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.4 | 8.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 10.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 4.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 5.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 4.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 3.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 0.8 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.2 | 1.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 25.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 14.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 18.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 10.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 8.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 1.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 18.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 5.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0034235 | GPI anchor binding(GO:0034235) metallodipeptidase activity(GO:0070573) |
| 0.1 | 6.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 24.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 17.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.5 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 18.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 4.9 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.6 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 7.0 | GO:0016787 | hydrolase activity(GO:0016787) |
| 0.0 | 3.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 11.4 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 9.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 8.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 113.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.9 | 83.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 79.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.4 | 25.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 16.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 29.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 13.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 16.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 35.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 8.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 9.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 29.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 4.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 117.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 4.7 | 79.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 2.0 | 51.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.5 | 78.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.5 | 105.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 1.2 | 20.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.1 | 48.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.8 | 41.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.6 | 31.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 28.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.4 | 22.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 10.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 7.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 4.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 30.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 4.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 2.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 8.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 12.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 7.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 4.9 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 9.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 3.8 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |