Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
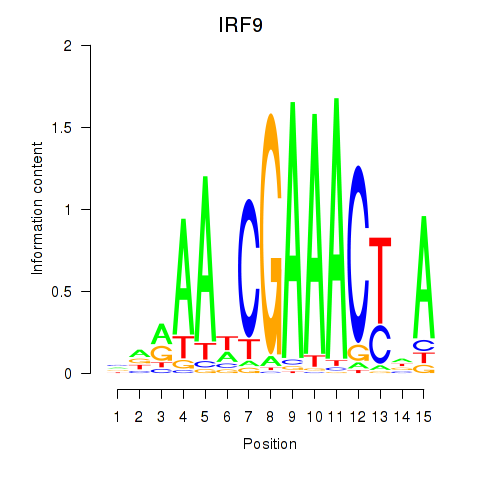
Results for IRF9
Z-value: 3.11
Transcription factors associated with IRF9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF9
|
ENSG00000213928.4 | interferon regulatory factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF9 | hg19_v2_chr14_+_24630465_24630531 | 0.75 | 1.3e-04 | Click! |
Activity profile of IRF9 motif
Sorted Z-values of IRF9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.7 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 2.3 | 9.0 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 1.5 | 31.5 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 1.3 | 15.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.2 | 3.6 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 1.2 | 4.7 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 1.1 | 10.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.7 | 8.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.7 | 2.0 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.5 | 1.0 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.5 | 62.1 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.4 | 1.2 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.3 | 2.4 | GO:0030578 | PML body organization(GO:0030578) |
| 0.3 | 1.5 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.3 | 2.7 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.2 | 1.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 0.6 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.2 | 0.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.1 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.1 | 0.4 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 1.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 1.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.5 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 2.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.2 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 3.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.9 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 1.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.0 | 0.3 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 2.9 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.3 | GO:2000809 | regulation of postsynaptic density protein 95 clustering(GO:1902897) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 7.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 2.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 1.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 2.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 3.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 28.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 6.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 27.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 8.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 32.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 35.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 1.2 | 4.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.9 | 11.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 10.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.4 | 4.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 1.2 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.3 | 8.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.3 | 2.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.6 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.2 | 0.7 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 3.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 4.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 11.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 31.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 9.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 3.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 2.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 9.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 115.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.7 | 2.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.7 | 11.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 4.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |