Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
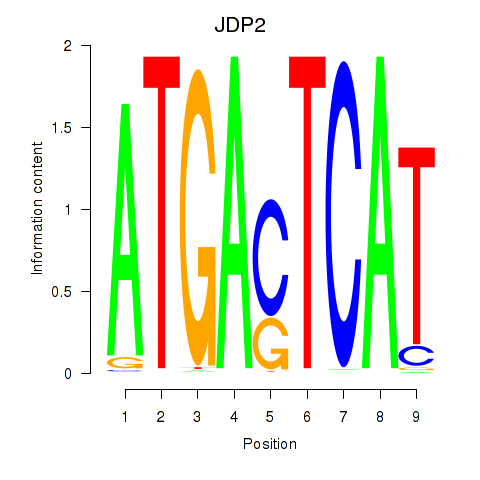
Results for JDP2
Z-value: 3.39
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JDP2 | hg19_v2_chr14_+_75894391_75894435 | 0.37 | 1.0e-01 | Click! |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 2.0 | 5.9 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 1.8 | 10.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 1.2 | 4.8 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 1.2 | 17.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 1.1 | 5.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.1 | 4.3 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 1.1 | 3.2 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 1.1 | 18.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.0 | 7.8 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.9 | 4.7 | GO:0052053 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.9 | 2.8 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.9 | 2.7 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.8 | 12.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.8 | 10.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.7 | 7.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.7 | 6.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.7 | 6.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 11.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.5 | 4.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.5 | 10.8 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.5 | 1.4 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.4 | 9.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 4.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.4 | 1.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.4 | 1.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 3.0 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 1.2 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 2.7 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 3.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 1.0 | GO:0052229 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.2 | 7.3 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.2 | 1.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 2.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 2.6 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 13.7 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 0.8 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.2 | 0.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 9.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 1.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 3.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 3.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 2.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 0.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 3.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.2 | 2.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 7.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 4.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 6.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 8.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 1.6 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.5 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.1 | 0.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.6 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 2.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 1.0 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 7.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.0 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.1 | 2.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.2 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 | 1.9 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 1.0 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 0.6 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.1 | 1.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 3.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 1.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.6 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.9 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.7 | GO:1990182 | exosomal secretion(GO:1990182) |
| 0.0 | 1.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 4.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 3.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.0 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 1.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 2.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.3 | GO:0061368 | negative regulation of growth hormone secretion(GO:0060125) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 1.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 2.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.4 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 7.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 1.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 1.5 | 13.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.1 | 11.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.7 | 9.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 7.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 18.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 3.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 1.5 | GO:0044753 | amphisome(GO:0044753) |
| 0.4 | 3.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 1.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 6.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 10.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.2 | 4.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.2 | 1.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 1.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.2 | 18.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.9 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 18.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 3.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 2.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 6.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 5.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 4.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 7.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 4.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 6.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 2.1 | 6.3 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 2.0 | 5.9 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 1.9 | 5.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.6 | 11.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.5 | 13.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.3 | 8.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.1 | 4.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.9 | 6.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.7 | 4.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.6 | 1.9 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.6 | 1.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 17.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 3.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 2.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.4 | 1.7 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.4 | 2.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 3.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 1.3 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.3 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 6.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 9.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 3.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 7.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 1.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 6.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.2 | 2.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 1.2 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 3.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 18.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.2 | 9.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 1.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 1.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.6 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 3.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 7.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 4.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.6 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 10.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 6.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 2.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 7.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 5.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 5.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 5.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 3.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.6 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 3.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 12.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 18.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 6.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 8.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 13.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 7.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 10.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 8.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 7.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 3.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 8.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 5.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 6.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 10.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 10.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 5.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.2 | 13.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 12.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 6.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 12.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 4.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 6.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 3.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 7.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 3.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 8.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 3.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |