Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
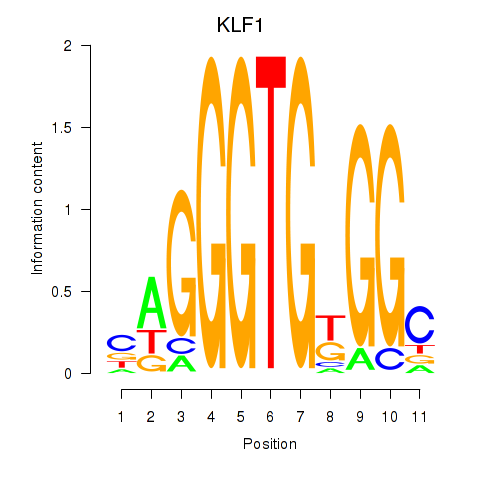
Results for KLF1
Z-value: 6.35
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | Kruppel like factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg19_v2_chr19_-_12997995_12998021 | -0.02 | 9.5e-01 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 4.0 | 12.1 | GO:0048627 | myoblast development(GO:0048627) |
| 3.6 | 21.6 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 3.3 | 20.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 2.6 | 7.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 2.6 | 20.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 2.4 | 14.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 2.2 | 30.5 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 2.1 | 10.7 | GO:1903284 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 2.0 | 5.9 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 1.8 | 7.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 1.7 | 8.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.7 | 10.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.7 | 27.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.7 | 6.7 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.6 | 29.6 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.6 | 9.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.6 | 10.9 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.6 | 9.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 1.5 | 17.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 1.5 | 4.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 1.4 | 62.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 1.4 | 9.9 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 1.4 | 9.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 1.3 | 7.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.2 | 12.0 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 1.2 | 21.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.2 | 11.8 | GO:0015820 | leucine transport(GO:0015820) |
| 1.2 | 3.5 | GO:0050904 | diapedesis(GO:0050904) |
| 1.2 | 3.5 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 1.2 | 15.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 1.1 | 5.5 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.1 | 18.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.0 | 4.2 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 1.0 | 16.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.0 | 10.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 1.0 | 5.9 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 1.0 | 3.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 1.0 | 17.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.9 | 3.6 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.9 | 27.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.9 | 6.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.8 | 7.6 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.8 | 9.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.8 | 7.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.8 | 13.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.8 | 10.6 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.7 | 3.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.7 | 3.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.7 | 3.6 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.7 | 0.7 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.7 | 4.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.7 | 4.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.7 | 38.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.7 | 10.6 | GO:0015816 | glycine transport(GO:0015816) |
| 0.7 | 2.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.6 | 3.1 | GO:0090649 | rRNA transport(GO:0051029) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.6 | 2.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.6 | 3.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.6 | 3.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.6 | 4.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.6 | 9.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.6 | 2.9 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.6 | 3.3 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.5 | 6.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.5 | 3.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.5 | 18.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 38.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.5 | 4.2 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.5 | 6.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.5 | 1.6 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.5 | 4.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.5 | 2.0 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.5 | 4.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.5 | 2.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.5 | 26.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.5 | 1.5 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.5 | 2.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.5 | 15.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.5 | 4.8 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.5 | 4.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 2.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.5 | 12.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 1.8 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.5 | 4.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.8 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.4 | 9.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 2.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 1.7 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.4 | 8.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 1.6 | GO:0033206 | meiotic chromosome movement towards spindle pole(GO:0016344) meiotic cytokinesis(GO:0033206) |
| 0.4 | 8.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 2.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.4 | 8.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.4 | 2.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.4 | 3.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.4 | 4.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.4 | 3.8 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.4 | 2.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.4 | 2.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 1.1 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.4 | 8.9 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.3 | 3.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 3.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 11.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.3 | 1.7 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.3 | 4.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 4.9 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.3 | 5.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 2.8 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.3 | 3.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 6.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 3.0 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.3 | 1.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 4.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 2.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.3 | 0.3 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.3 | 14.6 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.3 | 10.0 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.3 | 3.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 8.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 6.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 5.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 0.5 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 5.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 5.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 2.7 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.2 | 4.4 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.2 | 2.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 0.7 | GO:0090238 | negative regulation of icosanoid secretion(GO:0032304) regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 8.6 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.2 | 3.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 | 5.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.2 | 1.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 7.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.2 | 4.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 3.1 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 2.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 3.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.2 | 4.8 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 0.4 | GO:0071315 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.2 | 4.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 1.9 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.2 | 12.9 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 7.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.2 | 4.7 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.2 | 2.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 2.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.2 | 0.5 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.2 | 9.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 1.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 14.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 1.7 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 0.9 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 7.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 2.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 6.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 19.0 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 7.0 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 1.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 1.9 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.4 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 1.0 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 3.8 | GO:0010628 | positive regulation of gene expression(GO:0010628) |
| 0.1 | 1.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 5.3 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.1 | 1.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 3.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 3.8 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 1.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.3 | GO:0045553 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.1 | 2.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 1.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 3.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 3.2 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 3.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 2.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 7.1 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 1.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 7.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.4 | GO:0021707 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 2.8 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 3.9 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 1.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 2.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 1.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 3.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 2.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 3.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 4.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.3 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 0.2 | GO:0072254 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 4.5 | GO:0030888 | regulation of B cell proliferation(GO:0030888) |
| 0.1 | 1.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 2.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 2.0 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 1.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 2.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 5.4 | GO:0016032 | viral process(GO:0016032) multi-organism cellular process(GO:0044764) |
| 0.0 | 0.4 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 2.5 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 2.8 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.8 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.9 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 27.0 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 1.5 | 16.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.5 | 4.4 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 1.5 | 8.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.4 | 7.0 | GO:0071942 | XPC complex(GO:0071942) |
| 1.4 | 25.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.0 | 7.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.9 | 12.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.9 | 2.7 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.9 | 10.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 21.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.8 | 3.4 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.8 | 4.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.8 | 14.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.8 | 4.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.8 | 2.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.8 | 6.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.8 | 12.0 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.7 | 3.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.7 | 29.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.7 | 3.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.7 | 3.5 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.7 | 4.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.7 | 3.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.6 | 10.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.6 | 3.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 6.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.6 | 7.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 24.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 15.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.5 | 7.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 10.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.5 | 18.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.5 | 38.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 5.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.4 | 4.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 43.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 13.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 6.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 20.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 10.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 7.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 2.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 17.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 1.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 4.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 9.5 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 5.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.0 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.2 | 3.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 14.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 3.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 0.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 20.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 5.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 3.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 12.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 19.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 12.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 21.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 3.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 8.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 13.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 3.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 14.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 2.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 3.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 6.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 8.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 3.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 14.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 16.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 2.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 5.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 5.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 4.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 4.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 6.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 3.4 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 3.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 4.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 9.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 5.5 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 1.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 41.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 32.2 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 1.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 2.6 | 26.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.5 | 12.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 2.2 | 31.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 2.1 | 10.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.6 | 8.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 1.6 | 9.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.5 | 17.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.4 | 5.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 1.3 | 30.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.2 | 13.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.2 | 17.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.1 | 4.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 1.1 | 4.5 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.0 | 3.1 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 1.0 | 71.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 1.0 | 81.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.9 | 12.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.9 | 3.5 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.8 | 23.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.8 | 4.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.7 | 5.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.7 | 3.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.7 | 2.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.7 | 10.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.7 | 4.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.6 | 4.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.6 | 8.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.6 | 11.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 2.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 4.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.6 | 7.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.6 | 12.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.6 | 7.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.6 | 2.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.5 | 4.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.5 | 2.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 3.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.5 | 7.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 2.8 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.5 | 20.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.5 | 11.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 3.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.4 | 5.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.4 | 17.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 7.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 5.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 2.7 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 2.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 1.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.4 | 1.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.4 | 3.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 2.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.3 | 3.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.3 | 2.0 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 1.7 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 7.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 13.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 0.8 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.3 | 3.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 10.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 3.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 3.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 2.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 9.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 3.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 6.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 2.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 5.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 16.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 4.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 8.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 7.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 2.5 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 7.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.0 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 9.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 4.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 41.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 12.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 5.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 16.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 38.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 11.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 3.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 3.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 4.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 7.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 5.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 9.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 3.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 7.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 3.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 3.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 16.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 20.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 5.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 9.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.1 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.1 | 7.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 2.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 9.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 2.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 6.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 9.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 13.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 7.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 50.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 0.6 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 1.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 4.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 5.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 6.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 4.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.6 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 20.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.3 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 4.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.5 | GO:0051020 | GTPase binding(GO:0051020) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 43.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 60.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.5 | 30.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.5 | 12.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 9.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 5.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.3 | 77.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 16.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 9.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 11.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 7.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 16.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 23.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 13.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 12.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 9.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 8.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 5.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 4.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 5.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 6.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 7.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 8.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 10.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 8.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 4.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 10.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 21.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 5.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 17.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 31.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 17.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 15.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.6 | 21.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.4 | 11.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 18.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 18.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 18.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 6.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.4 | 6.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 2.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.3 | 16.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 12.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 10.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 8.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 3.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 29.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.3 | 17.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 4.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 6.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 1.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 26.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 8.6 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.2 | 11.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 1.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 6.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 7.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 10.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 6.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 11.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 6.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 6.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 3.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 18.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 9.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 3.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.9 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 5.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 30.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 3.3 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 3.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 5.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 5.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |