Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
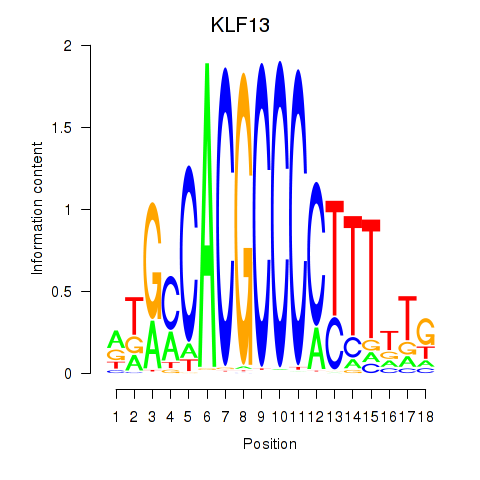
Results for KLF13
Z-value: 1.41
Transcription factors associated with KLF13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF13
|
ENSG00000169926.5 | Kruppel like factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF13 | hg19_v2_chr15_+_31619013_31619095 | -0.48 | 3.2e-02 | Click! |
Activity profile of KLF13 motif
Sorted Z-values of KLF13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 1.1 | 3.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.8 | 2.3 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.4 | 1.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.4 | 1.4 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.3 | 1.6 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.3 | 5.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 1.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.3 | 1.1 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.3 | 1.0 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.2 | 0.7 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 | 1.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.2 | 0.6 | GO:0048003 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 1.3 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.2 | 1.6 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 0.8 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 2.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 2.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.8 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.6 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 1.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.1 | 4.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.7 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 1.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.5 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.9 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 2.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.3 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 1.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 1.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.6 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 1.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.6 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 1.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.3 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 2.4 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 2.0 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 2.8 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 2.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.3 | 5.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.6 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 2.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.4 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 4.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.4 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 1.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 2.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.0 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 2.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.5 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.7 | 3.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.4 | 1.6 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.4 | 1.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 1.3 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.3 | 0.8 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.2 | 1.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 6.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 0.7 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.2 | 2.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.5 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.2 | 1.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 2.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.6 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 2.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 2.3 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 4.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 5.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 6.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 4.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 2.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.9 | REACTOME MEIOSIS | Genes involved in Meiosis |