Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
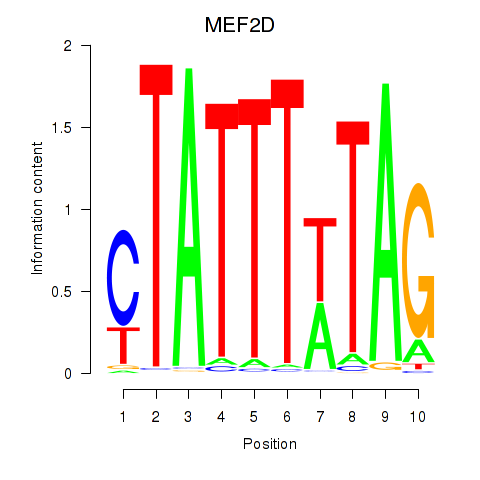
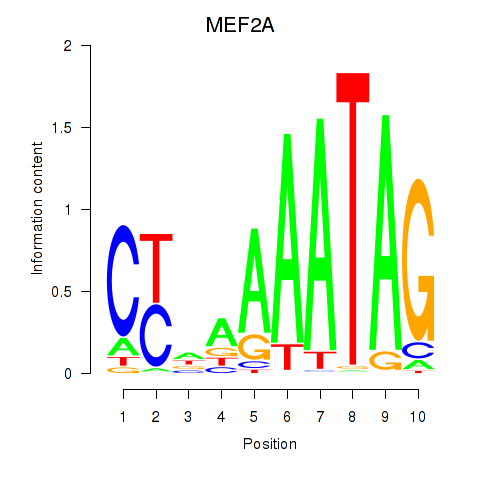
Results for MEF2D_MEF2A
Z-value: 2.47
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2D | hg19_v2_chr1_-_156470515_156470542 | -0.65 | 1.9e-03 | Click! |
| MEF2A | hg19_v2_chr15_+_100106244_100106292 | 0.21 | 3.7e-01 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 41.6 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 3.0 | 9.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.3 | 12.9 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.1 | 3.3 | GO:1903217 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 1.0 | 5.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 1.0 | 5.8 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.0 | 3.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.9 | 2.7 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.9 | 7.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.8 | 2.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 | 2.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.6 | 1.7 | GO:1904640 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.5 | 3.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 2.6 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.5 | 3.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.4 | 10.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.4 | 1.7 | GO:0018277 | protein deamination(GO:0018277) |
| 0.4 | 1.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.4 | 2.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 2.0 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.4 | 3.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 2.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 1.5 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) |
| 0.4 | 2.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 1.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 4.4 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.3 | 1.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.3 | 2.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 9.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.3 | 1.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 0.9 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.3 | 0.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 2.4 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 5.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 0.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 1.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.3 | 3.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 3.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 3.0 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.3 | 0.5 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.2 | 1.4 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.2 | 3.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 2.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.8 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.2 | 2.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.8 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.2 | 0.8 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.2 | 0.8 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 3.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 1.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 3.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.7 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.2 | 6.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 0.9 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.3 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.2 | 0.9 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 5.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 1.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.7 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.7 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.8 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 3.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 4.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 5.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.5 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 2.0 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 3.4 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 1.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.7 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 4.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 3.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 1.1 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.1 | 2.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.7 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.4 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 2.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.7 | GO:0061615 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 3.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 2.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.4 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 7.8 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.1 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 3.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 4.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.8 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.4 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.9 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 2.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 7.3 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.8 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 2.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 1.3 | GO:0018212 | peptidyl-tyrosine phosphorylation(GO:0018108) peptidyl-tyrosine modification(GO:0018212) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.5 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 2.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 1.8 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 1.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.5 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 1.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.7 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 1.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.8 | 2.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 3.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.6 | 2.5 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.6 | 2.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 5.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 5.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 3.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 4.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 2.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 1.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 2.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 5.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.8 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 3.0 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.9 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 2.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 7.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.3 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 3.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 3.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 4.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 30.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 3.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 2.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 10.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.9 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 5.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 5.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 8.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 4.4 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 10.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 1.1 | 3.3 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 1.0 | 9.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.8 | 4.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.6 | 5.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.6 | 9.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.6 | 1.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.6 | 1.7 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.5 | 1.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.5 | 3.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 2.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 6.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 2.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 1.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 49.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.4 | 7.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 12.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 1.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.4 | 3.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.5 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.3 | 4.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 1.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 3.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 9.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 1.1 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.3 | 1.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 5.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 1.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 1.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.8 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.2 | 5.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.0 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 2.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 1.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.2 | 1.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 1.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.2 | 4.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 3.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 2.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 1.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 5.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.9 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 2.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 1.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 3.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.4 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.5 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 3.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 1.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 4.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.9 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 1.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 4.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 3.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 3.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 4.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 39.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 12.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 7.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 2.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 4.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 5.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 5.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 5.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 37.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.5 | 9.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 5.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 10.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 3.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 6.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 5.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 7.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 6.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 9.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 3.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 5.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 3.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 6.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 4.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |