Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
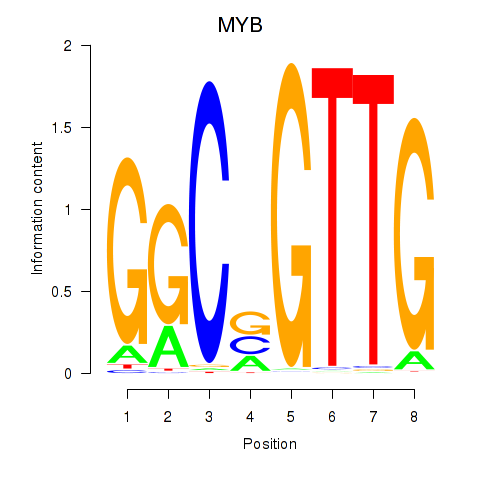
Results for MYB
Z-value: 4.37
Transcription factors associated with MYB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYB
|
ENSG00000118513.14 | MYB proto-oncogene, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYB | hg19_v2_chr6_+_135502466_135502489 | 0.85 | 1.8e-06 | Click! |
Activity profile of MYB motif
Sorted Z-values of MYB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 11.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 1.7 | 18.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 1.4 | 5.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.3 | 6.3 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 1.1 | 9.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.0 | 2.0 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 1.0 | 11.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.0 | 2.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.9 | 3.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.8 | 2.5 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.8 | 2.5 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.8 | 2.3 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.8 | 2.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 | 2.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.7 | 2.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.7 | 6.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.7 | 2.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.7 | 0.7 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.7 | 4.7 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.7 | 3.3 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.6 | 1.9 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.6 | 1.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.6 | 1.9 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.6 | 4.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.6 | 4.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.6 | 5.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.5 | 5.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.5 | 0.5 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.5 | 6.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.5 | 3.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.5 | 1.0 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.5 | 2.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.5 | 3.9 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.5 | 1.9 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.5 | 1.4 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.5 | 3.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.5 | 2.4 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.5 | 0.9 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.5 | 9.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.5 | 1.8 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.5 | 3.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.4 | 1.8 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.4 | 3.5 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 4.9 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.4 | 1.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.4 | 2.5 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.4 | 4.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 1.2 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.4 | 1.5 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.4 | 1.8 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.3 | 1.0 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.3 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.3 | 3.7 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 1.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.7 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 1.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 1.6 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 5.8 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 3.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 1.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.3 | 1.6 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.3 | 1.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 1.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.3 | 1.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 | 8.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.3 | 1.7 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 0.8 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.3 | 1.1 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.3 | 1.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 | 6.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 0.8 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.3 | 4.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 1.6 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.3 | 3.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 1.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.3 | 1.3 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.3 | 1.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 4.0 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.3 | 1.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.3 | 2.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 6.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 4.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.3 | 0.3 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.3 | 1.0 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 3.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 3.9 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.2 | 1.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 2.2 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.2 | 0.9 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.2 | 1.9 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.2 | 0.7 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 1.6 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 1.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.2 | 2.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 4.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 1.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.5 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 2.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.9 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.2 | 0.6 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 9.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 2.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 0.6 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.2 | 1.6 | GO:0060136 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.6 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.2 | 0.4 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.2 | 2.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 7.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.9 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 8.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 1.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.2 | GO:0046666 | retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.2 | 1.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.2 | 0.5 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.9 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.2 | 1.3 | GO:0036102 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.2 | 1.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 0.4 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.2 | 0.5 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 3.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 1.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.2 | 1.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 9.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.2 | 0.8 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 4.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 1.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 0.6 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 5.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.2 | 3.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 1.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 1.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.9 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 3.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.6 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 1.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 18.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 | 0.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 5.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.7 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 3.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:0060319 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 1.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.8 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 2.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 3.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.5 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 1.0 | GO:0043570 | meiotic mismatch repair(GO:0000710) maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 3.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 1.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.7 | GO:0000387 | spliceosomal tri-snRNP complex assembly(GO:0000244) spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.8 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 2.6 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.1 | 2.7 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.1 | 1.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 3.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.8 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 3.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.6 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 1.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.9 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.1 | 1.0 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 1.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.2 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.1 | 0.3 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.7 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 1.9 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.6 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 2.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.9 | GO:0015867 | ADP transport(GO:0015866) ATP transport(GO:0015867) |
| 0.1 | 1.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 4.0 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.1 | 2.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 2.0 | GO:0061387 | regulation of axon extension(GO:0030516) regulation of extent of cell growth(GO:0061387) |
| 0.1 | 2.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 1.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 2.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 1.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 2.6 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.1 | 0.5 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 3.9 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.3 | GO:0061304 | cerebellum vasculature development(GO:0061300) retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 4.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.6 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 2.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 5.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.9 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.1 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.2 | GO:0006090 | pyruvate metabolic process(GO:0006090) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 7.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 2.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 2.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.8 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.9 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.5 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.3 | GO:0010903 | negative regulation of cytokine secretion involved in immune response(GO:0002740) negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 1.0 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 1.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.5 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.8 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 2.1 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 0.2 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 2.0 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.5 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.1 | 0.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.8 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 2.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.7 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.5 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.6 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.8 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 1.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.5 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.8 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 1.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.1 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 1.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 3.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 1.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0090209 | negative regulation of triglyceride biosynthetic process(GO:0010868) negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 2.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 2.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.5 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 1.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.6 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 1.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.5 | GO:0046461 | neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 4.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.3 | GO:0045844 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 | 0.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 6.4 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 1.8 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 3.4 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.7 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 2.0 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.8 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.7 | GO:0099536 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) synaptic signaling(GO:0099536) trans-synaptic signaling(GO:0099537) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.7 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.0 | GO:1904029 | regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 2.0 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.5 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 3.4 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 2.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 3.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.6 | GO:0045595 | regulation of cell differentiation(GO:0045595) |
| 0.0 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 2.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 2.5 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.3 | GO:0010648 | negative regulation of cell communication(GO:0010648) negative regulation of signaling(GO:0023057) |
| 0.0 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0060346 | sequestering of TGFbeta in extracellular matrix(GO:0035583) bone trabecula formation(GO:0060346) |
| 0.0 | 1.0 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 1.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.1 | GO:0061571 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 1.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 1.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.7 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.4 | GO:0043065 | positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.4 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.0 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 1.9 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.1 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.9 | 5.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.9 | 11.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.9 | 6.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.8 | 2.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 2.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.7 | 2.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.7 | 2.8 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.6 | 4.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 2.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.6 | 2.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.5 | 1.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.5 | 5.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 1.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 1.7 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.4 | 2.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 1.7 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.4 | 2.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 1.6 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.4 | 2.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 1.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 1.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.4 | 1.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.4 | 2.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 3.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 1.4 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.3 | 1.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 9.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 4.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 13.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 3.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 1.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 19.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 0.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 2.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 2.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 2.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.1 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 4.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 0.8 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 8.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 3.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 7.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.7 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 1.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 8.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 6.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 3.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 2.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 2.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 2.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 2.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 4.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 6.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 1.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 1.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 16.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 5.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.5 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 1.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 6.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.8 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 4.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 11.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 2.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.2 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.1 | 1.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 4.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 4.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 7.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.1 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 3.0 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 2.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 5.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.5 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 3.0 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.7 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 4.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 1.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 2.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 8.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0042588 | zymogen granule(GO:0042588) zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 5.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.5 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 1.0 | 4.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.9 | 7.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.9 | 6.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.9 | 2.6 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.8 | 2.5 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.8 | 2.3 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.7 | 2.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.7 | 6.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 4.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.6 | 2.6 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.6 | 1.9 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.6 | 1.9 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.6 | 3.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.6 | 1.9 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.6 | 2.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.5 | 1.6 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.5 | 3.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.5 | 2.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.5 | 1.8 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 1.7 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.4 | 1.3 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.4 | 3.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.4 | 1.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 1.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.4 | 1.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 3.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 1.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.3 | 1.3 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.3 | 1.0 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.3 | 1.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.3 | 1.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.3 | 3.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 2.1 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.3 | 9.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.3 | 0.8 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 2.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 0.8 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.3 | 1.6 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.3 | 3.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 7.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 12.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 2.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 1.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.2 | 2.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 1.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 3.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 0.7 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.2 | 1.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 14.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 0.6 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.2 | 0.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 2.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 0.9 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 0.7 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 1.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.5 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 0.9 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.2 | 0.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.2 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.5 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 2.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 3.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.6 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.2 | 2.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 2.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 1.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 6.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.9 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 2.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.3 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 3.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 2.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 4.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.7 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 3.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 1.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.8 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 5.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.3 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 3.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 4.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 2.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 1.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.3 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 8.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 1.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 3.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 3.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 4.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 7.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 2.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0032405 | mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 10.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 4.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 2.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 4.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0005138 | ciliary neurotrophic factor receptor binding(GO:0005127) interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 4.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 9.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.7 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 1.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 5.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 24.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 18.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 12.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 11.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 11.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 3.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 6.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 5.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 3.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 3.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 3.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 3.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.3 | 12.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 9.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 12.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.3 | 5.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 7.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 3.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 2.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 27.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 1.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 2.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 3.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 8.8 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 1.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 9.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 4.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 5.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 2.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 2.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 2.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 2.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 3.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 5.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 3.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |