Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
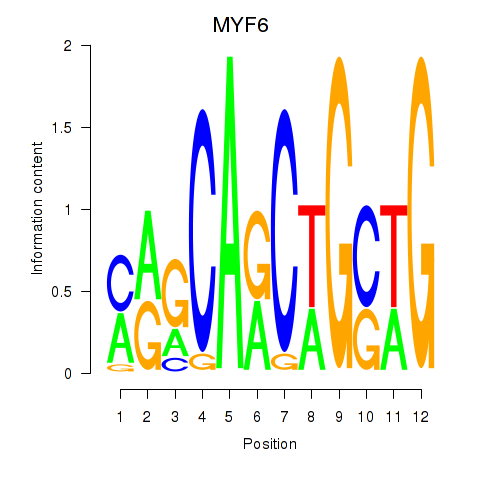
Results for MYF6
Z-value: 4.58
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.3 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYF6 | hg19_v2_chr12_+_81101277_81101321 | -0.34 | 1.4e-01 | Click! |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 4.4 | 17.5 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 3.7 | 14.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 2.1 | 8.3 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 1.6 | 7.9 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 1.6 | 3.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.5 | 17.5 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 1.4 | 4.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 1.4 | 8.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.3 | 6.6 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.3 | 5.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.2 | 3.7 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 1.2 | 8.6 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 1.2 | 3.6 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 1.2 | 4.6 | GO:0003335 | corneocyte development(GO:0003335) |
| 1.2 | 3.5 | GO:0035284 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 1.1 | 3.3 | GO:0035989 | tendon development(GO:0035989) |
| 1.0 | 4.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.0 | 4.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 1.0 | 10.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 1.0 | 9.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 1.0 | 3.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 1.0 | 5.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.0 | 3.0 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.9 | 2.8 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.9 | 4.6 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.9 | 6.4 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.9 | 8.9 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.9 | 3.6 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.8 | 5.1 | GO:0072069 | ascending thin limb development(GO:0072021) DCT cell differentiation(GO:0072069) metanephric ascending thin limb development(GO:0072218) metanephric DCT cell differentiation(GO:0072240) |
| 0.8 | 10.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.8 | 2.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.8 | 3.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 3.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.8 | 5.5 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.8 | 3.2 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.8 | 3.2 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.8 | 15.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.8 | 4.7 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.8 | 3.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.8 | 11.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.8 | 5.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.7 | 3.0 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.7 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.7 | 9.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.7 | 10.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.7 | 3.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.7 | 2.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.7 | 2.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.6 | 3.9 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.6 | 12.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.6 | 5.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.6 | 1.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.6 | 9.0 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 1.8 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 2.9 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.6 | 3.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.6 | 2.3 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.6 | 4.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.5 | 3.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.5 | 3.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.5 | 9.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.5 | 2.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.5 | 1.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.5 | 3.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.5 | 2.9 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.5 | 4.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.5 | 0.9 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.5 | 3.2 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.5 | 5.9 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.5 | 1.4 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.4 | 9.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.4 | 2.6 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.4 | 1.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.4 | 1.7 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.4 | 3.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 1.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.4 | 1.2 | GO:1900369 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.4 | 3.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.4 | 5.6 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.4 | 7.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.4 | 0.4 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.4 | 2.2 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.4 | 1.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.4 | 2.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.4 | 1.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.4 | 2.5 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.4 | 1.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 4.5 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.3 | 1.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 2.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 1.3 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.3 | 5.9 | GO:0030238 | male sex determination(GO:0030238) |
| 0.3 | 2.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.3 | 0.3 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.3 | 4.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.3 | 4.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.3 | 1.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.3 | 2.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 1.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.3 | 0.9 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.3 | 7.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 1.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.3 | 1.4 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.3 | 1.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 3.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 4.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 2.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 3.0 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.3 | 9.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.3 | 0.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 2.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.3 | 1.6 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 1.0 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.3 | 4.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 0.8 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.3 | 1.0 | GO:0005997 | xylulose metabolic process(GO:0005997) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 11.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 0.7 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.2 | 3.1 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 0.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 1.8 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 5.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.2 | GO:2000981 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.2 | 1.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 0.9 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 1.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 0.2 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.2 | 1.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 27.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.2 | 1.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.2 | 3.7 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.2 | 4.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 2.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 0.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 8.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 4.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.2 | 0.8 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 1.4 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 1.7 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.2 | 0.8 | GO:1902532 | negative regulation of intracellular signal transduction(GO:1902532) |
| 0.2 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 1.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.2 | 6.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 4.0 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.2 | 3.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 3.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 1.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 4.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.7 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.2 | 1.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 3.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 2.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 6.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 1.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.2 | 3.7 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 | 4.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.6 | GO:1902430 | negative regulation of beta-amyloid formation(GO:1902430) |
| 0.2 | 0.2 | GO:0002339 | B cell selection(GO:0002339) |
| 0.2 | 0.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 0.5 | GO:0051714 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.2 | 1.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 4.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 1.5 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.2 | 2.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 1.5 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) insulin metabolic process(GO:1901142) |
| 0.2 | 3.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 1.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 14.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 4.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.5 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.1 | 2.7 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 1.6 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.7 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 1.0 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 3.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 1.7 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.7 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 2.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 2.6 | GO:0048562 | embryonic organ morphogenesis(GO:0048562) |
| 0.1 | 2.8 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 0.4 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.1 | 1.3 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 1.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.3 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 2.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.6 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.1 | 0.8 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.8 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 1.3 | GO:0021527 | ventral spinal cord interneuron differentiation(GO:0021514) spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 2.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 3.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.9 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 1.3 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.5 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.1 | 2.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.6 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 2.8 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 1.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.4 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 1.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 3.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 3.3 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 3.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.6 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.4 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 1.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 3.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.2 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.1 | 4.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 1.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.5 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.1 | 0.3 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.2 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.1 | 8.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.9 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 1.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.4 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.7 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 8.9 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 0.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.9 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 1.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.5 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.0 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 2.4 | GO:0009607 | response to biotic stimulus(GO:0009607) |
| 0.1 | 0.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 1.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.9 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 2.9 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.1 | 2.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.0 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.6 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 1.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.2 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.1 | 0.3 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 1.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 0.8 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 0.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 3.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 7.1 | GO:0050880 | regulation of blood vessel size(GO:0050880) |
| 0.1 | 4.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 1.9 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 3.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 4.3 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0046851 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 | 1.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 2.0 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.9 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.7 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.4 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 1.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 2.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.9 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 0.3 | GO:2000352 | negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 | 0.6 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.6 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 1.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.7 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0051459 | regulation of corticotropin secretion(GO:0051459) cellular response to cocaine(GO:0071314) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 4.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 1.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.5 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 2.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.5 | GO:0097050 | type B pancreatic cell apoptotic process(GO:0097050) regulation of type B pancreatic cell apoptotic process(GO:2000674) |
| 0.0 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 2.5 | GO:0006511 | ubiquitin-dependent protein catabolic process(GO:0006511) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 1.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 1.2 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.0 | GO:0021772 | olfactory bulb development(GO:0021772) |
| 0.0 | 2.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) initiation of neural tube closure(GO:0021993) |
| 0.0 | 2.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.9 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 4.0 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:0023061 | signal release(GO:0023061) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 2.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 1.2 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 2.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 2.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 3.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 1.5 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 2.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.9 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.6 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.5 | GO:0006974 | cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 1.5 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.3 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.1 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.3 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.3 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.5 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.7 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 16.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.4 | 4.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.3 | 3.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 1.2 | 9.2 | GO:1990393 | 3M complex(GO:1990393) |
| 1.1 | 3.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.0 | 5.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.7 | 3.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.7 | 3.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.7 | 7.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.7 | 2.7 | GO:0060187 | cell pole(GO:0060187) |
| 0.7 | 2.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.6 | 14.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 3.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.6 | 15.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 3.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.5 | 15.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.5 | 2.5 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.5 | 2.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.5 | 2.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 4.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 4.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 2.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 5.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.4 | 1.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.4 | 2.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 3.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.4 | 1.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 2.5 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 2.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 4.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 5.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 3.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 1.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 4.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) chromaffin granule(GO:0042583) |
| 0.2 | 4.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.7 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 3.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 3.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 3.7 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.2 | 2.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 1.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.2 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.6 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.2 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 2.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 3.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 1.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 2.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 14.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 2.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.0 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 4.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 6.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 9.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 13.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 2.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 25.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 14.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 5.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 4.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 4.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 2.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.8 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.7 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 4.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 5.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 4.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.5 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 5.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 28.7 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 5.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 7.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 2.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 2.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 1.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 4.1 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 5.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 8.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 10.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 3.7 | 14.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 3.5 | 17.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 2.9 | 8.6 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 2.3 | 9.2 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 2.1 | 4.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 2.0 | 2.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 1.5 | 4.6 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.4 | 5.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.4 | 4.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 1.4 | 4.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.3 | 3.9 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.3 | 3.9 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 1.2 | 10.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.2 | 3.6 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 1.2 | 3.6 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 1.2 | 14.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 1.1 | 3.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 1.1 | 14.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 1.0 | 3.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.9 | 4.7 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.9 | 2.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.9 | 5.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.9 | 4.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.8 | 2.4 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.8 | 3.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.8 | 2.3 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.8 | 3.9 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.8 | 9.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.7 | 2.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.7 | 15.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 2.2 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.7 | 3.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.7 | 4.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.7 | 2.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.6 | 4.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.6 | 12.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.6 | 5.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.5 | 12.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 2.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.5 | 8.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.5 | 4.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.5 | 2.9 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.5 | 1.8 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.5 | 5.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.4 | 1.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 4.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.4 | 9.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.4 | 17.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 5.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 1.2 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.4 | 1.2 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.4 | 7.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 3.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 11.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 3.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 3.6 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.4 | 11.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 1.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.4 | 6.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 1.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.4 | 1.8 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.4 | 5.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 1.4 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.4 | 15.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 11.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 3.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 2.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.3 | 2.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.3 | 1.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.3 | 1.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 9.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 3.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 3.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 1.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.3 | 20.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 1.6 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 1.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.3 | 2.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 2.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 2.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.3 | 2.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 2.7 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 5.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 2.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.9 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 2.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 7.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 5.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 10.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 0.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.8 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.2 | 0.6 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 0.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 3.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 4.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.2 | 1.5 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 0.7 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.2 | 4.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 3.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 1.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 1.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 5.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.3 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.2 | 0.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 18.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.2 | 3.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 0.5 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 6.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 14.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 3.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 2.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 4.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 4.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.9 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 4.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 5.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 1.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.9 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.3 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 3.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 6.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 4.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 3.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 2.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 11.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 4.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 2.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.2 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 1.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 7.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 4.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 1.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 5.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 1.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 4.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 8.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 2.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 4.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.5 | GO:0050656 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 5.3 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 2.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 2.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 4.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 1.0 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 3.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.6 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 2.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 6.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.9 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 1.1 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 1.2 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 25.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 2.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 11.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 16.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 13.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 1.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 5.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 11.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 14.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 15.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 6.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 4.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 5.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 7.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 3.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 5.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 3.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 3.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 4.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 7.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 8.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 6.5 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 23.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.5 | 15.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 7.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 20.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.4 | 11.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 7.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 5.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 3.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 5.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 3.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 14.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 9.9 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.3 | 8.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 12.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 6.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 4.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 19.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.2 | 3.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 1.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.2 | 3.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 7.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 1.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 2.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 2.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 4.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 16.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 3.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 10.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 17.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 6.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 5.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 6.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 9.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 8.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 5.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 3.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.9 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 3.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 2.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 6.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 5.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 7.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 1.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 1.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |