Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
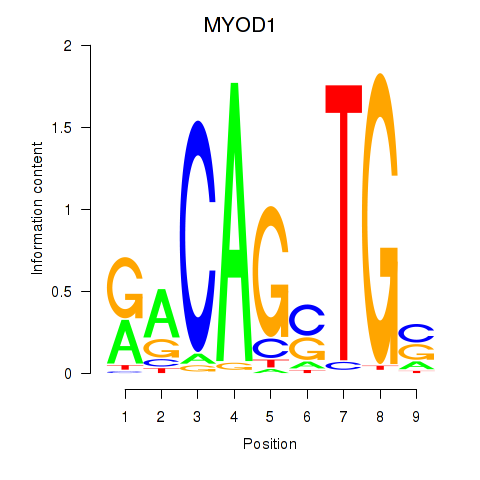
Results for MYOD1
Z-value: 1.84
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.3 | myogenic differentiation 1 |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.9 | 5.6 | GO:0018032 | protein amidation(GO:0018032) |
| 0.9 | 16.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.8 | 3.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.8 | 0.8 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.7 | 2.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.7 | 10.1 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.7 | 2.8 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.7 | 2.0 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.6 | 3.2 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.5 | 1.6 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.5 | 1.6 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.5 | 0.5 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.5 | 1.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.5 | 1.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.5 | 2.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 2.2 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.4 | 1.7 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.4 | 1.6 | GO:1904501 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.4 | 1.9 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.4 | 1.9 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 9.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 2.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.4 | 4.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 0.7 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.4 | 2.8 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.4 | 1.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 2.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 | 2.7 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.3 | 1.4 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.3 | 3.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 4.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.3 | 1.3 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) sensory system development(GO:0048880) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.3 | 1.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 2.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 6.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 6.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 0.9 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.3 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.3 | 0.9 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 0.8 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 2.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.3 | 1.9 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 1.6 | GO:0060437 | lung growth(GO:0060437) |
| 0.3 | 0.8 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.3 | 1.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 0.8 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.3 | 1.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 2.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 4.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 8.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 | 1.0 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 0.6 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 1.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 3.5 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 0.6 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.2 | 0.8 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 1.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.2 | 1.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 1.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 2.7 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 3.1 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 1.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 0.8 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.1 | 0.4 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 1.8 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.5 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 1.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 2.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.5 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.1 | 1.9 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.4 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 3.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.6 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.8 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 0.9 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.3 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 1.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 3.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 1.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 4.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.4 | GO:0070997 | neuron death(GO:0070997) |
| 0.1 | 1.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.6 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.3 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.4 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 1.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 2.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 2.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 1.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.7 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 2.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.9 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 6.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 2.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 1.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 1.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.4 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 1.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.3 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 2.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 1.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.3 | GO:0014870 | response to muscle inactivity(GO:0014870) |
| 0.1 | 0.5 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 1.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 1.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.3 | GO:1901675 | response to methylglyoxal(GO:0051595) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 5.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.0 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 1.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.2 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 1.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.5 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 0.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.1 | 1.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 1.7 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 0.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 2.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.7 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 1.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 1.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 2.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 1.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 1.0 | GO:0070977 | bone maturation(GO:0070977) |
| 0.0 | 0.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 3.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.5 | GO:0070884 | calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) L-cystine transport(GO:0015811) |
| 0.0 | 0.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.5 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.9 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.5 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.8 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 2.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 1.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 2.2 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 1.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 2.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 2.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.0 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.0 | GO:0055025 | positive regulation of cardiac muscle tissue development(GO:0055025) |
| 0.0 | 2.2 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:1902961 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 3.8 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 2.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.3 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 2.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.6 | GO:0045445 | myoblast differentiation(GO:0045445) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.5 | 2.6 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.5 | 1.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 3.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 1.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 1.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 1.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.6 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 3.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 0.8 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 2.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 0.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 5.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 3.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 3.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 4.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 2.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 4.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 2.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 5.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 1.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 6.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 5.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 4.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 4.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 2.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 11.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 13.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 2.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 2.8 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 3.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 2.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 5.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.1 | GO:0070603 | SWI/SNF superfamily-type complex(GO:0070603) |
| 0.0 | 1.7 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 16.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.9 | 5.6 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.6 | 1.9 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.6 | 4.5 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 1.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 2.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 1.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 10.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.4 | 8.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.4 | 1.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.4 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.3 | 2.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 4.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 2.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 1.2 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.3 | 0.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 0.9 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 0.8 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 0.8 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.2 | 2.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 2.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 0.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 0.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 6.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 1.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 4.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 2.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.9 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 1.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 5.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.3 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 1.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 2.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 3.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 6.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 4.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.7 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.8 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 2.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 3.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.7 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 7.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 4.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 1.6 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 2.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0015193 | L-cystine transmembrane transporter activity(GO:0015184) L-glutamine transmembrane transporter activity(GO:0015186) L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 2.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 3.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 4.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 2.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 11.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 7.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 5.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 4.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 7.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 4.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 5.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.4 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 2.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 7.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 6.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 2.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.7 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 4.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 1.0 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.3 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |