Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
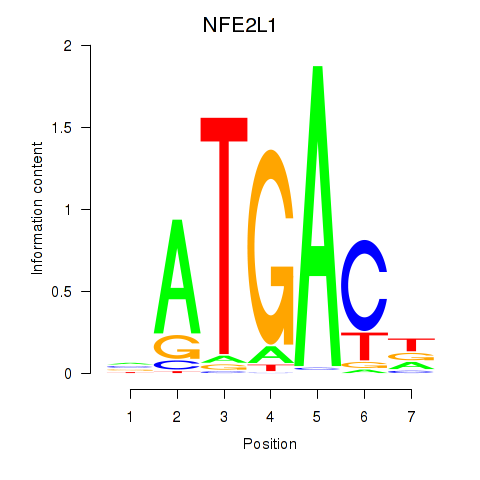
Results for NFE2L1
Z-value: 2.63
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | nuclear factor, erythroid 2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg19_v2_chr17_+_46125707_46125746 | 0.91 | 3.0e-08 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 19.9 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 2.3 | 27.8 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 2.3 | 6.9 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 2.2 | 15.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.6 | 4.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.5 | 16.6 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 1.4 | 4.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 1.4 | 5.4 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.2 | 3.6 | GO:2000283 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.1 | 12.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.0 | 6.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.0 | 5.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.9 | 2.8 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.9 | 2.8 | GO:0002580 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.9 | 2.8 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.8 | 4.2 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.7 | 2.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.7 | 2.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.7 | 4.1 | GO:0042335 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.7 | 3.3 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.6 | 3.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.5 | 3.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 1.8 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 3.5 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.4 | 1.3 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.4 | 1.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.4 | 4.3 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.4 | 2.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 1.4 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.4 | 2.5 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.3 | 1.3 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 | 2.6 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.3 | 3.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.3 | 2.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 4.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 0.9 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.3 | 1.9 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 4.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 1.3 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.2 | 2.0 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 2.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 1.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 1.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.2 | 2.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.9 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 0.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.8 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.2 | 2.6 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.8 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.2 | 1.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.8 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.9 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 2.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 3.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.2 | 0.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 2.0 | GO:2001181 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.6 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.2 | 0.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 3.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 1.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 6.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.5 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.7 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 1.8 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 1.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 5.4 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 2.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 5.5 | GO:0010575 | positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.1 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 1.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.1 | 18.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 0.9 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.5 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 1.6 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 1.0 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.9 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 2.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.6 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 1.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 3.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.4 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 2.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 2.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.7 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 1.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 0.7 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.5 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 1.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 3.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.6 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 2.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.4 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 2.5 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 4.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.5 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 1.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 2.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 2.7 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 4.8 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 3.8 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 1.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.3 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 1.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.6 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.8 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.9 | GO:0097205 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.9 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.6 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 1.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.7 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 1.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.6 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 1.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 3.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 1.9 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.5 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 1.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 1.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.1 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.0 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 11.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.3 | GO:0046649 | lymphocyte activation(GO:0046649) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 2.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0097459 | ferric iron import(GO:0033216) iron ion import into cell(GO:0097459) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) iron ion import across plasma membrane(GO:0098711) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.3 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.6 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.1 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.4 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 2.5 | 19.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.9 | 2.8 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.8 | 4.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.8 | 4.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.7 | 4.3 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 2.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.5 | 1.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.5 | 1.8 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 4.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.4 | 3.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 1.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.3 | 3.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 5.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 5.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 17.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 5.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 5.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 2.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.6 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 4.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 18.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 5.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 3.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 4.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 3.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 4.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 11.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 3.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 15.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 2.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 3.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 15.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.2 | 3.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 1.0 | 4.0 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.9 | 12.0 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.8 | 4.2 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.7 | 4.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.6 | 6.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 5.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 13.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.5 | 2.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.5 | 18.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 2.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 16.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.4 | 2.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 1.9 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.4 | 1.9 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.4 | 3.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 2.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.3 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 4.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 0.9 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 0.9 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 0.9 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.2 | 1.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.2 | 0.8 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 3.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 18.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 5.7 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.2 | 2.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 1.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 3.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 3.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 4.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 3.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 1.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 2.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 18.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 3.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 4.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 5.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 2.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 4.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.5 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 3.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 22.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.5 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.5 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 2.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 1.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 1.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 7.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 1.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 3.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 27.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 17.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 8.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 17.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 5.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 9.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 6.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 4.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 3.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 3.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 3.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 18.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 7.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.3 | 2.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 5.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 3.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 3.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 6.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 4.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 17.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 6.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 3.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.5 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 1.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 15.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 5.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 3.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 3.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 3.1 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 7.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |