Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
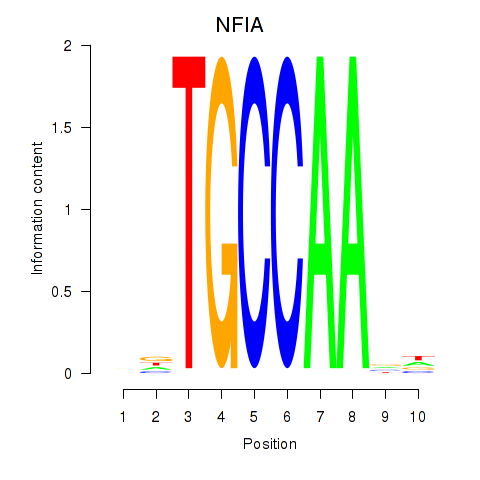
Results for NFIA
Z-value: 3.40
Transcription factors associated with NFIA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIA
|
ENSG00000162599.11 | nuclear factor I A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIA | hg19_v2_chr1_+_61330931_61331017 | 0.92 | 1.4e-08 | Click! |
Activity profile of NFIA motif
Sorted Z-values of NFIA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 4.0 | 12.0 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 2.7 | 8.0 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 1.8 | 5.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.8 | 8.8 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.4 | 5.6 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 1.2 | 3.6 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 1.1 | 8.0 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 1.1 | 6.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.1 | 20.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.0 | 4.0 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 1.0 | 12.0 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 1.0 | 2.9 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 1.0 | 5.9 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.9 | 3.4 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.8 | 2.5 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.8 | 28.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.8 | 6.8 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.7 | 6.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.7 | 1.5 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.7 | 15.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.7 | 2.8 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.7 | 3.3 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.7 | 4.6 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.6 | 1.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.6 | 3.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.6 | 5.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.6 | 3.9 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.5 | 9.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.5 | 8.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.5 | 2.4 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.5 | 4.4 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.5 | 3.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.5 | 3.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.5 | 1.9 | GO:0035905 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.5 | 6.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.5 | 0.9 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.4 | 3.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.4 | 14.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.4 | 6.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.4 | 0.9 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.4 | 4.9 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.4 | 21.7 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.4 | 2.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 1.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.4 | 1.6 | GO:1990927 | short-term synaptic potentiation(GO:1990926) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.4 | 1.5 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.4 | 2.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.4 | 1.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 3.8 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.3 | 2.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 2.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 2.6 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.3 | 1.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.3 | 3.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.3 | 4.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 1.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 2.4 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 2.4 | GO:0030578 | PML body organization(GO:0030578) |
| 0.3 | 3.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.3 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 1.6 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.3 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.3 | 3.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 1.6 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 6.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.3 | 1.5 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 5.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 1.4 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.2 | 2.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 2.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 0.9 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 2.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 2.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 5.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 1.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 1.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 9.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 4.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 5.2 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.2 | 1.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.7 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.2 | 1.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 2.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 0.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.2 | 0.7 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 4.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 3.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 4.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 8.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 0.8 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.6 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 1.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 3.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.5 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.1 | 0.7 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 3.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 1.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.0 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 1.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.7 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.1 | 2.9 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.6 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 1.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 3.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 3.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 2.5 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.1 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.1 | 1.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 6.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 2.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 3.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.9 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.5 | GO:0051168 | nuclear export(GO:0051168) |
| 0.1 | 4.9 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 19.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 9.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 22.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 1.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 2.9 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.4 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 3.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.8 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.6 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.1 | 4.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.8 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 0.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.3 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 1.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 1.0 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 12.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.6 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.6 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 1.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 1.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 9.9 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 1.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 2.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0055065 | metal ion homeostasis(GO:0055065) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.8 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.9 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 1.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.8 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 1.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 1.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.7 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 1.1 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 1.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 2.3 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.7 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 3.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 3.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.0 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 1.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 3.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 1.0 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 1.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 2.4 | GO:1903052 | positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) |
| 0.0 | 0.3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.6 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 3.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 1.4 | 8.1 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 6.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 46.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 8.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.4 | 3.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.4 | 4.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 1.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 1.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.3 | 24.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 1.6 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.3 | 1.2 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.3 | 1.2 | GO:0070939 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.3 | 2.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 1.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 2.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 4.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 3.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 1.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.3 | 2.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 4.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 1.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 5.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 2.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 5.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 0.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 2.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 1.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 3.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 1.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 13.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 2.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 10.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.4 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 6.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 9.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 19.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 2.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 16.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 9.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 1.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 3.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 2.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 2.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 5.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 5.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 4.6 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 1.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 6.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 3.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 3.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 3.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.9 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.9 | 5.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.3 | 3.9 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 1.1 | 4.6 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.9 | 3.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.9 | 2.6 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.8 | 2.5 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.8 | 18.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.8 | 4.9 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.7 | 2.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.6 | 5.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.6 | 8.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 2.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.5 | 2.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 1.6 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 1.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.4 | 2.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 3.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.9 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.4 | 4.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.3 | 2.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 2.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 47.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 14.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 5.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 6.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 11.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 3.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 2.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 3.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 3.9 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 1.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 7.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 9.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 1.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 2.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 6.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 0.7 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.2 | 8.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 24.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.2 | 10.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.9 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 0.8 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 4.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 4.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 4.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 3.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.6 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 1.3 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 3.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 6.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 3.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 3.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.4 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 5.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 6.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 3.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.7 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 3.2 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 1.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 5.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 3.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 2.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.1 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.3 | GO:0031751 | D2 dopamine receptor binding(GO:0031749) D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 2.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 12.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 10.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.5 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 1.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 3.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 4.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.9 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.0 | GO:0030674 | protein binding, bridging(GO:0030674) binding, bridging(GO:0060090) |
| 0.1 | 1.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 8.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 1.8 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 2.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 3.7 | GO:0044212 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 1.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 31.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 5.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 2.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 14.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 2.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 11.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 39.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 16.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 10.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 7.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 15.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 16.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 4.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 10.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 35.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 8.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 9.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 4.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 5.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 6.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 14.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 4.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 4.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 8.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 4.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 5.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 6.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 10.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 5.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 6.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 5.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 3.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 4.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 13.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 1.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 7.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 4.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 3.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.6 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.1 | 4.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 6.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 10.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 8.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 3.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 3.7 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 1.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 2.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |