Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
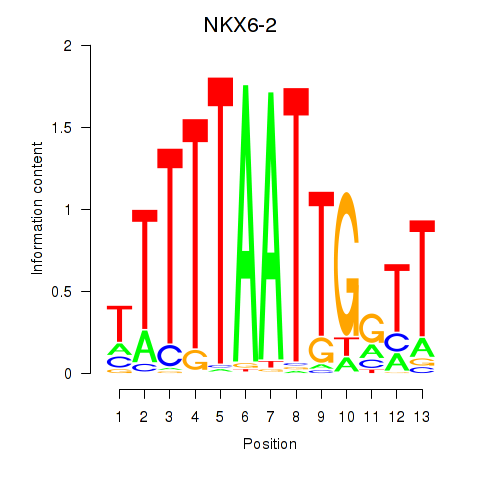
Results for NKX6-2
Z-value: 1.22
Transcription factors associated with NKX6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-2
|
ENSG00000148826.6 | NK6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX6-2 | hg19_v2_chr10_-_134599556_134599653 | -0.17 | 4.8e-01 | Click! |
Activity profile of NKX6-2 motif
Sorted Z-values of NKX6-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.4 | 2.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.3 | 1.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 1.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.9 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 3.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.2 | 0.5 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 1.8 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.8 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.1 | 3.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.7 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 2.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.5 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 1.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 3.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.7 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.1 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.1 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.4 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.6 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 6.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.2 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.6 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:0046469 | plasma lipoprotein particle oxidation(GO:0034441) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0070863 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.3 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 2.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.6 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 3.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 5.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 4.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 2.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.8 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.1 | 4.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.3 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 4.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 2.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 4.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |