Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
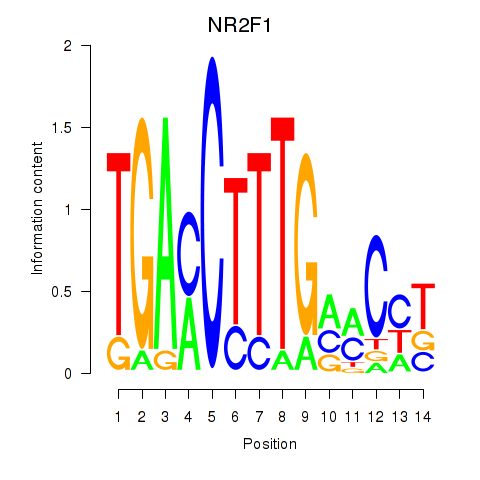
Results for NR2F1
Z-value: 2.31
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | nuclear receptor subfamily 2 group F member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F1 | hg19_v2_chr5_+_92919043_92919082 | 0.90 | 5.2e-08 | Click! |
Activity profile of NR2F1 motif
Sorted Z-values of NR2F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.8 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 3.1 | 9.2 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 1.9 | 5.6 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 1.5 | 4.4 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 1.4 | 4.1 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 1.3 | 3.8 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.3 | 12.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 1.2 | 2.5 | GO:0097187 | dentinogenesis(GO:0097187) |
| 1.0 | 3.0 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.0 | 2.9 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.9 | 2.6 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.9 | 9.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.8 | 3.3 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.8 | 3.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.7 | 6.9 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.6 | 2.5 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.6 | 2.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.5 | 1.6 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.5 | 2.3 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.4 | 3.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 3.5 | GO:0046618 | drug export(GO:0046618) |
| 0.4 | 1.5 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.4 | 1.4 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.3 | 1.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 4.0 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.3 | 1.9 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.3 | 1.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.3 | 2.0 | GO:0042262 | DNA protection(GO:0042262) |
| 0.3 | 6.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 1.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.3 | 1.3 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 1.0 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 2.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.7 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 2.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 6.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 4.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.6 | GO:1904640 | response to methionine(GO:1904640) |
| 0.2 | 1.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 3.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 1.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 1.5 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 1.9 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 1.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.5 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.2 | 0.8 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.2 | 3.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 1.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.7 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 5.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.7 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.1 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 1.5 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 1.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.5 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.7 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 2.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 3.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 2.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.6 | GO:1905040 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 1.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:0035922 | pulmonary valve formation(GO:0003193) visceral motor neuron differentiation(GO:0021524) foramen ovale closure(GO:0035922) |
| 0.1 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.3 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.2 | GO:2000532 | renal protein absorption(GO:0097017) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.1 | 0.4 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 1.3 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 0.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.8 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 2.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 3.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.2 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.1 | 5.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 1.4 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 2.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 2.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 7.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.3 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.9 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.5 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.4 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.5 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 3.1 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 1.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.2 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 3.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 1.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 4.4 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 0.5 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.3 | GO:0060770 | positive thymic T cell selection(GO:0045059) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 1.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.3 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.2 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 1.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.4 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0002861 | regulation of inflammatory response to antigenic stimulus(GO:0002861) negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 1.0 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.4 | 1.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 3.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 3.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 4.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 3.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.7 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 1.5 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 5.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 2.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 4.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 9.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 5.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 2.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 5.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 1.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 1.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 6.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 7.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 9.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 1.3 | 3.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.0 | 3.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.9 | 2.7 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.8 | 4.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.8 | 3.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.7 | 7.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.7 | 4.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.6 | 3.5 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.6 | 12.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.5 | 9.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.5 | 1.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.5 | 2.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 3.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.3 | 1.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 4.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.2 | 2.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 4.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.2 | 4.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.0 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.2 | 0.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 1.0 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.2 | 1.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 0.8 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 0.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 1.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.6 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.2 | 0.5 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 0.7 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.5 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 4.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 3.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 1.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 3.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 4.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 2.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 5.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 1.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 2.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.9 | GO:0052658 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 1.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.3 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 2.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.6 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 2.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 6.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 3.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 11.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 13.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 12.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 3.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 5.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 4.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 4.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 3.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 5.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 3.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 3.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 4.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 3.0 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 2.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |