Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
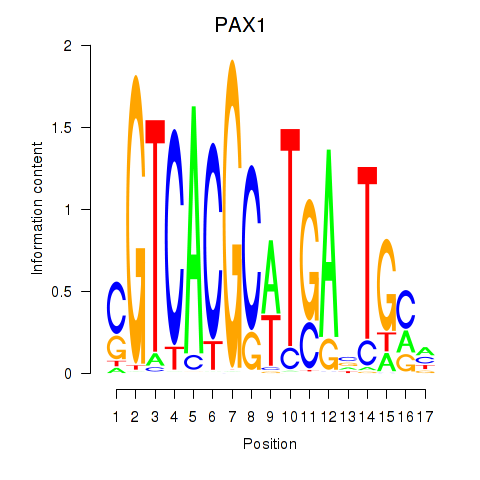
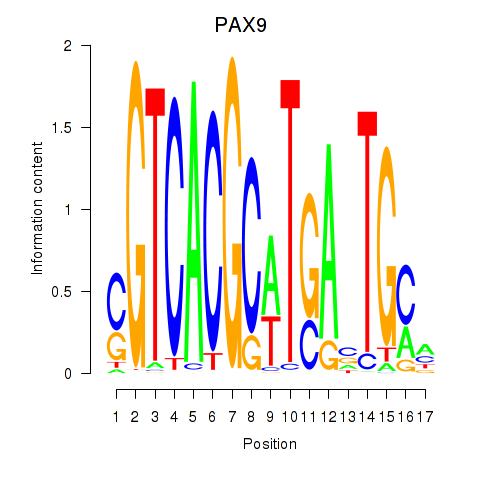
Results for PAX1_PAX9
Z-value: 1.38
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX1
|
ENSG00000125813.9 | paired box 1 |
|
PAX9
|
ENSG00000198807.8 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg19_v2_chr14_+_37131058_37131139 | -0.92 | 6.7e-09 | Click! |
| PAX1 | hg19_v2_chr20_+_21686290_21686311 | -0.05 | 8.5e-01 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 1.4 | 5.8 | GO:0018277 | protein deamination(GO:0018277) |
| 0.6 | 1.8 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.3 | 2.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 0.8 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 0.6 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.2 | 1.9 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.9 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.5 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.7 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 1.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 2.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 2.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 7.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 1.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 1.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 5.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 2.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.6 | 2.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 7.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 0.6 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.2 | 1.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.2 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 2.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 2.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |