Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
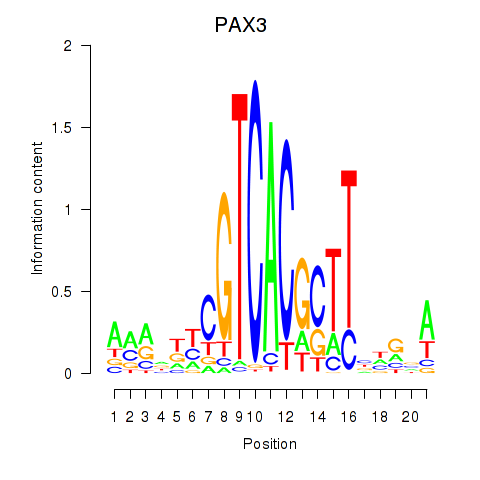
Results for PAX3
Z-value: 0.86
Transcription factors associated with PAX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX3
|
ENSG00000135903.14 | paired box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX3 | hg19_v2_chr2_-_223163465_223163730 | -0.45 | 4.6e-02 | Click! |
Activity profile of PAX3 motif
Sorted Z-values of PAX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.6 | 3.0 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.3 | 2.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 1.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 0.6 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.2 | 0.9 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 0.9 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.2 | 0.9 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 1.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.3 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.3 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.3 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.1 | 0.3 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.6 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.9 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.6 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.3 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 1.8 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 1.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.5 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.3 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 2.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.9 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 1.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 1.2 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.2 | 0.7 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.6 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.6 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 0.3 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 2.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 2.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 2.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 3.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 3.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 2.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 3.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |