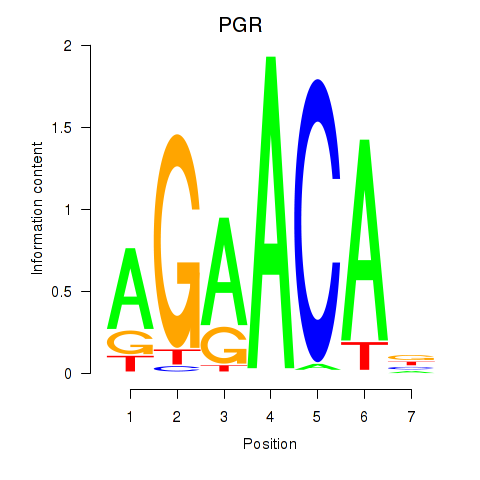
| 0.5 |
1.5 |
GO:2001190 |
positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 |
0.4 |
GO:0060734 |
regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.1 |
0.4 |
GO:0002728 |
negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 |
0.5 |
GO:0035482 |
gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.1 |
0.5 |
GO:0002384 |
hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.1 |
0.3 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 |
0.5 |
GO:0090362 |
positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 |
0.3 |
GO:0070253 |
somatostatin secretion(GO:0070253) |
| 0.1 |
0.4 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
0.4 |
GO:2000110 |
negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 |
0.3 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.1 |
0.4 |
GO:0018032 |
protein amidation(GO:0018032) |
| 0.1 |
0.3 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.1 |
0.2 |
GO:1904237 |
regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 |
0.2 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.2 |
GO:1904428 |
negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 |
0.3 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 |
0.9 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
1.1 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
0.2 |
GO:0042412 |
taurine biosynthetic process(GO:0042412) |
| 0.0 |
0.1 |
GO:1901207 |
regulation of heart looping(GO:1901207) |
| 0.0 |
0.2 |
GO:1990523 |
bone regeneration(GO:1990523) |
| 0.0 |
0.1 |
GO:0048691 |
modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 |
0.2 |
GO:0051510 |
regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 |
0.2 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.2 |
GO:1903284 |
negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 |
0.1 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 |
0.4 |
GO:0070294 |
renal sodium ion absorption(GO:0070294) |
| 0.0 |
0.4 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 |
0.1 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 |
0.5 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.0 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.1 |
GO:0072615 |
nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 |
0.7 |
GO:0006704 |
glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 |
0.1 |
GO:0018057 |
peptidyl-lysine oxidation(GO:0018057) |
| 0.0 |
0.1 |
GO:0043049 |
hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 |
0.3 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.1 |
GO:0090214 |
spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 |
0.2 |
GO:1900625 |
regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 |
0.9 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 |
0.1 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 |
0.1 |
GO:0030241 |
skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 |
0.1 |
GO:1903625 |
negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 |
0.1 |
GO:0009236 |
cobalamin biosynthetic process(GO:0009236) |
| 0.0 |
0.2 |
GO:0014718 |
positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 |
0.1 |
GO:1900226 |
negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 |
0.2 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.1 |
GO:0021571 |
rhombomere 5 development(GO:0021571) |
| 0.0 |
0.1 |
GO:0008057 |
eye pigment granule organization(GO:0008057) |
| 0.0 |
0.4 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:0030185 |
nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 |
0.3 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.7 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.1 |
GO:0042418 |
epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.0 |
0.4 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.1 |
GO:0051466 |
corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 |
0.2 |
GO:2000660 |
negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 |
0.1 |
GO:2000909 |
regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 |
0.0 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 |
0.4 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.4 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.1 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.1 |
GO:2000342 |
negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 |
0.1 |
GO:1900247 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 |
0.1 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.2 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.1 |
GO:0021557 |
optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 |
0.2 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.1 |
GO:0070444 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.2 |
GO:0052697 |
flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 |
0.0 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 |
0.4 |
GO:0070262 |
peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 |
0.1 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.0 |
0.2 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.0 |
0.3 |
GO:0090026 |
positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 |
0.1 |
GO:1900019 |
regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 |
0.1 |
GO:0071469 |
cellular response to alkaline pH(GO:0071469) |
| 0.0 |
0.2 |
GO:0099612 |
protein localization to axon(GO:0099612) |
| 0.0 |
0.1 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 |
0.0 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 |
0.0 |
GO:0001812 |
positive regulation of type I hypersensitivity(GO:0001812) B cell cytokine production(GO:0002368) |
| 0.0 |
0.1 |
GO:0042335 |
cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.0 |
0.3 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0046549 |
retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.2 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.1 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 |
0.1 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 |
0.3 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.1 |
GO:0060332 |
positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 |
0.1 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 |
0.1 |
GO:0009744 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |