Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
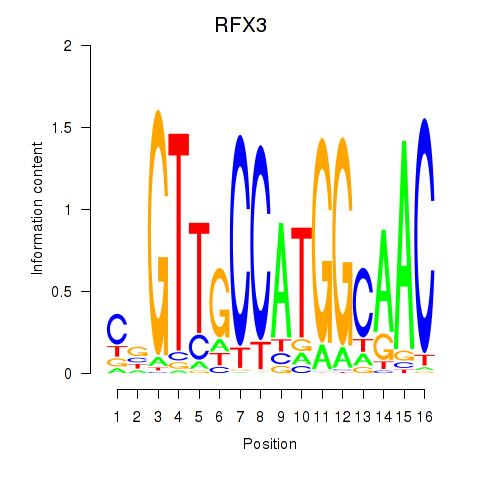
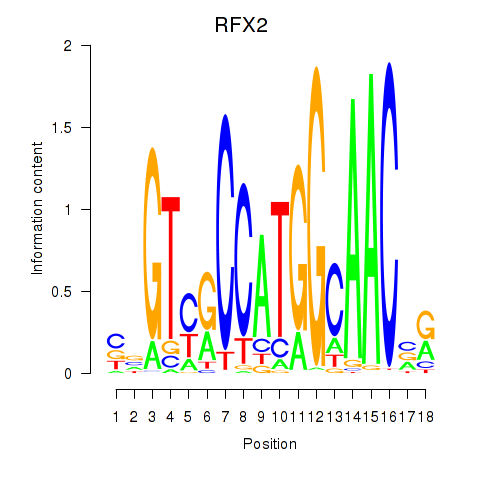
Results for RFX3_RFX2
Z-value: 4.61
Transcription factors associated with RFX3_RFX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX3
|
ENSG00000080298.11 | regulatory factor X3 |
|
RFX2
|
ENSG00000087903.8 | regulatory factor X2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX2 | hg19_v2_chr19_-_6110474_6110551 | 0.69 | 7.0e-04 | Click! |
| RFX3 | hg19_v2_chr9_-_3525968_3526016 | -0.46 | 4.0e-02 | Click! |
Activity profile of RFX3_RFX2 motif
Sorted Z-values of RFX3_RFX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX3_RFX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 2.2 | 6.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 2.0 | 5.9 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 1.8 | 7.3 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.7 | 5.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.7 | 5.0 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 1.5 | 4.6 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 1.3 | 5.3 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 1.3 | 5.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 1.2 | 3.7 | GO:0070121 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 1.1 | 8.0 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 1.1 | 3.3 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.1 | 12.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.1 | 9.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 1.1 | 5.4 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 1.1 | 4.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 1.0 | 5.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.9 | 2.7 | GO:0090340 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.9 | 4.5 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.8 | 9.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.8 | 2.4 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.8 | 3.2 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.8 | 11.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.7 | 2.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.7 | 3.4 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.7 | 5.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.7 | 2.7 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.6 | 1.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.6 | 1.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.6 | 3.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.6 | 2.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.6 | 12.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.6 | 2.4 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.6 | 2.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.6 | 0.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.6 | 19.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.6 | 5.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.6 | 2.8 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.6 | 3.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 5.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.5 | 2.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.5 | 1.9 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.4 | 11.5 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 7.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 4.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.4 | 3.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.4 | 2.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.4 | 1.1 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.4 | 3.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 1.9 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 7.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 4.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 2.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 2.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 2.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 8.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 2.3 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.2 | 4.0 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 2.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.2 | 1.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 1.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.6 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.2 | 1.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.2 | 1.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 3.7 | GO:1901978 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.2 | 1.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 8.7 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 1.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 4.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.2 | 0.6 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.2 | 2.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 0.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 7.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 5.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 0.9 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 1.7 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 1.3 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 | 2.8 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 0.3 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 2.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 1.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 9.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 3.3 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 2.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 2.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.9 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.1 | 1.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 7.3 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 1.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 2.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 3.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 2.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.4 | GO:0061571 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 1.2 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.1 | 2.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.8 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 10.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 4.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 5.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.7 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 3.9 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 2.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.5 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.5 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.1 | 15.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 1.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 5.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.6 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.2 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.1 | 1.3 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 2.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 2.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 4.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 1.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 2.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.1 | 0.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 4.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.3 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 5.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 2.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.6 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 2.2 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.1 | 2.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.7 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 2.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.3 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.1 | 2.7 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.1 | 0.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 4.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 2.3 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.1 | 0.9 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 6.9 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 3.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 3.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 5.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 0.2 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 4.1 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.2 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 7.4 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 3.5 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 2.5 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 3.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 1.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.6 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 1.8 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.6 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.4 | GO:0031344 | regulation of cell projection organization(GO:0031344) |
| 0.0 | 0.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0048670 | regulation of collateral sprouting(GO:0048670) positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 4.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 1.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.0 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 1.4 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 2.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.6 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 8.7 | GO:0050851 | antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 1.4 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 3.3 | GO:0071426 | ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.0 | 0.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 2.1 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.0 | 2.4 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.5 | GO:0001975 | response to amphetamine(GO:0001975) |
| 0.0 | 1.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.4 | GO:0009749 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.1 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.8 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.8 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.1 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 3.4 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 1.7 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.2 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.3 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.2 | 3.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 1.2 | 7.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.1 | 4.6 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.9 | 19.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.9 | 7.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.7 | 6.6 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.7 | 11.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 9.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.5 | 1.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 1.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.4 | 7.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 1.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.4 | 3.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 7.1 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 1.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.3 | 4.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 4.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 4.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 5.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 4.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 1.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 5.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 3.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 5.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 3.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.0 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.2 | 6.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 1.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 5.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 4.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 1.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 4.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 5.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 10.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 4.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 3.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 11.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 3.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 3.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 8.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 9.9 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 0.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 18.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 4.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 3.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) nuclear stress granule(GO:0097165) |
| 0.0 | 1.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 6.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 3.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 4.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 6.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 7.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 12.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 3.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.5 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 1.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.3 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 1.5 | 9.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.5 | 4.5 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 1.3 | 6.7 | GO:0002046 | opsin binding(GO:0002046) |
| 1.0 | 3.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.9 | 5.4 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 2.7 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.8 | 3.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.8 | 7.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.7 | 3.6 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.7 | 7.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.7 | 2.8 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.7 | 2.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.7 | 5.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 2.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.6 | 3.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.5 | 6.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.5 | 1.9 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.5 | 13.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 1.8 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.4 | 2.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 3.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.4 | 1.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 12.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 5.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.3 | 1.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 4.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 0.3 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.3 | 6.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 6.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 3.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 4.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 2.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 1.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 0.6 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 0.6 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.2 | 1.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 0.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 3.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 3.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 4.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 2.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.2 | 3.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 3.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 2.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.2 | 0.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 3.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 0.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 0.6 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.2 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 5.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 7.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 4.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.0 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.1 | 0.4 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 4.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 2.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 16.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 3.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 4.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 5.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 3.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 4.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 10.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.5 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 3.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 4.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 2.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 17.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 3.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 4.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.3 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 5.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 2.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 4.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 11.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 1.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 4.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 6.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 4.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 2.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 11.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 4.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 5.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.1 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 9.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 11.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 15.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 11.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 11.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 8.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 3.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.5 | 5.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 13.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 3.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 4.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 4.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 10.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 6.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 7.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 17.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 2.9 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 9.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 4.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 7.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 5.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 3.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.2 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 2.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 4.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 7.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 6.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.3 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 4.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 2.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |