Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
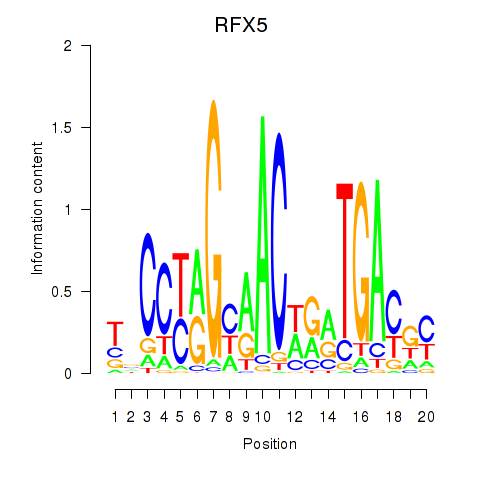
Results for RFX5
Z-value: 0.86
Transcription factors associated with RFX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX5
|
ENSG00000143390.13 | regulatory factor X5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX5 | hg19_v2_chr1_-_151319318_151319497 | -0.83 | 5.8e-06 | Click! |
Activity profile of RFX5 motif
Sorted Z-values of RFX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.9 | 3.4 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 | 0.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.7 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 3.9 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 3.3 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 2.2 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 3.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 3.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 2.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 3.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.7 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 3.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 3.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.2 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 2.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0052658 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 3.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |