Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
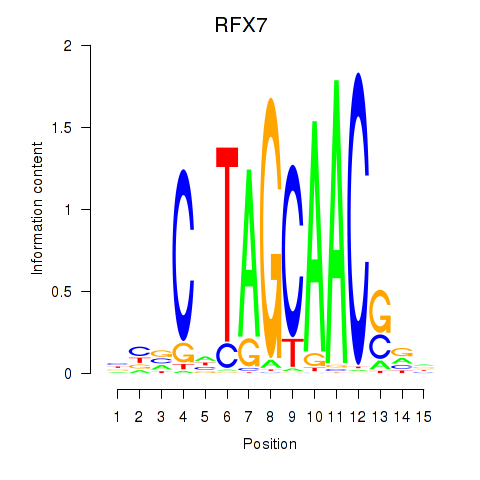
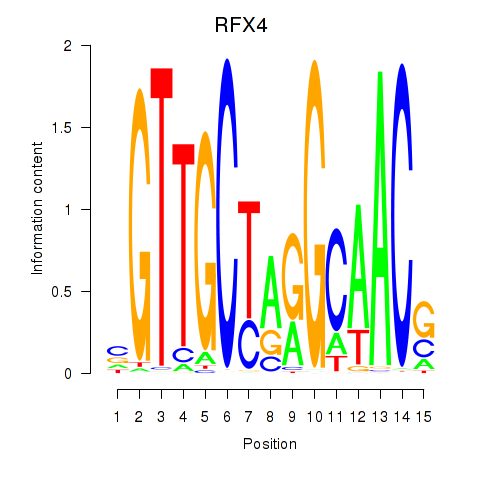
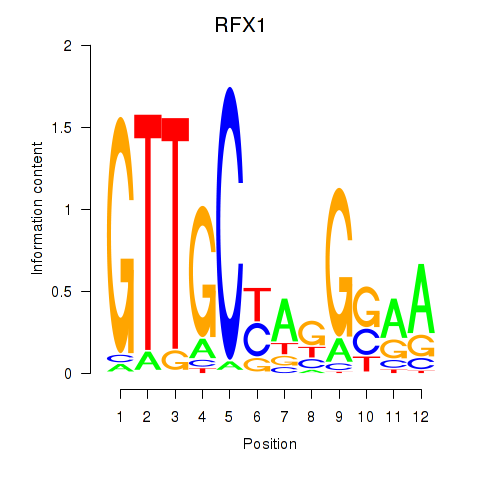
Results for RFX7_RFX4_RFX1
Z-value: 3.70
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX7
|
ENSG00000181827.10 | regulatory factor X7 |
|
RFX4
|
ENSG00000111783.8 | regulatory factor X4 |
|
RFX1
|
ENSG00000132005.4 | regulatory factor X1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | -0.83 | 5.1e-06 | Click! |
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | 0.74 | 1.7e-04 | Click! |
| RFX4 | hg19_v2_chr12_+_106994905_106994954 | 0.42 | 6.4e-02 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 34.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 2.7 | 8.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.9 | 5.8 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 1.2 | 3.6 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.1 | 4.3 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) |
| 1.0 | 4.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 4.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.7 | 2.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.7 | 7.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.7 | 3.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.6 | 1.8 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.6 | 7.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.6 | 4.5 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.5 | 1.6 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.5 | 5.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.5 | 2.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.5 | 1.4 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.4 | 1.8 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.4 | 1.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 3.0 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.4 | 1.5 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.4 | 1.4 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.4 | 2.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.3 | 0.7 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 6.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 1.0 | GO:0039022 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.3 | 1.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 1.2 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.3 | 1.4 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.3 | 4.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 4.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.3 | 1.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 1.6 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.3 | 2.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 0.8 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 2.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 0.2 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.2 | 1.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.0 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 7.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.7 | GO:0090108 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.2 | 0.7 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.2 | 0.2 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.2 | 1.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.2 | 0.6 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 2.6 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 | 0.4 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.2 | 1.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 2.9 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 4.6 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.2 | 0.6 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 1.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 1.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 1.6 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.2 | 1.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 2.7 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 1.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 0.6 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.2 | 2.3 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 6.5 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.2 | 1.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.2 | 0.5 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.2 | 0.5 | GO:0006106 | fumarate metabolic process(GO:0006106) glycerol biosynthetic process(GO:0006114) aspartate catabolic process(GO:0006533) |
| 0.2 | 2.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.5 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.2 | 0.5 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 0.8 | GO:0007228 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 | 0.5 | GO:0006186 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.2 | 1.7 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 0.8 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.2 | 0.5 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 0.5 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 70.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 6.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 1.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 1.3 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.1 | 1.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.9 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 20.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.5 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.4 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.1 | 0.5 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.1 | 8.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.0 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 0.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.7 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 2.5 | GO:0043485 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 2.0 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 2.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.7 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 2.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.4 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 0.6 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.6 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.9 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.3 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.1 | 3.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 2.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.3 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 1.0 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 | 0.2 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 1.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 10.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.2 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 1.7 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.1 | 1.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 2.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.3 | GO:0051958 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 0.5 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.6 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.8 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 3.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 1.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.9 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 2.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 5.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 2.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 2.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 1.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.7 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 1.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 1.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 1.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 3.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.5 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 2.4 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.4 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 2.3 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.2 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epinephrine secretion(GO:0048242) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.6 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 9.1 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 2.1 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.3 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 1.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0031344 | regulation of cell projection organization(GO:0031344) |
| 0.0 | 0.9 | GO:0042339 | keratan sulfate biosynthetic process(GO:0018146) keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 1.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 6.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.3 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 2.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 2.9 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 2.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 1.1 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.3 | GO:0006818 | hydrogen transport(GO:0006818) proton transport(GO:0015992) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 1.3 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.7 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.3 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.7 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 12.0 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.8 | 8.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.5 | 2.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.4 | 1.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 3.6 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.4 | 1.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 4.3 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 0.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 0.8 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.3 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 2.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 5.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 1.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 3.3 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.2 | 2.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 3.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 0.5 | GO:0001534 | radial spoke(GO:0001534) |
| 0.2 | 2.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 3.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 1.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 5.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 4.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 3.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 17.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.6 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.2 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.1 | 4.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 8.6 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.0 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 6.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 4.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.2 | GO:0016020 | membrane(GO:0016020) |
| 0.0 | 5.7 | GO:0042995 | cell projection(GO:0042995) |
| 0.0 | 2.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 3.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.1 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 3.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 6.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.8 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 1.2 | 3.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.1 | 4.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.9 | 2.8 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.9 | 2.8 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.8 | 7.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.7 | 6.3 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.7 | 0.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.6 | 2.3 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.5 | 4.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.5 | 3.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.5 | 67.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.4 | 1.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.4 | 3.6 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.4 | 1.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.4 | 8.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.4 | 1.5 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.4 | 8.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 1.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 0.9 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.3 | 1.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 2.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 5.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 1.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.2 | 1.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 3.5 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.2 | 0.7 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.2 | 2.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.6 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.2 | 0.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 0.6 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.2 | 1.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.5 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 0.8 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.2 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 1.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 1.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 0.5 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 5.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 2.7 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 0.5 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 11.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 3.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.9 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.7 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 1.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 9.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 14.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 6.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 3.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.5 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 2.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 2.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.9 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.5 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 1.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 1.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 12.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.7 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 3.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 3.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 2.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 1.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 11.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 2.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.3 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 3.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 4.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 3.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 3.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |