Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
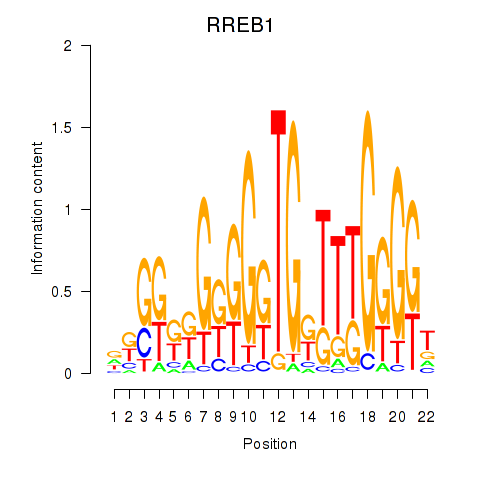
Results for RREB1
Z-value: 3.44
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7107999_7108054 | 0.95 | 7.3e-11 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 2.0 | 7.8 | GO:0007403 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 1.7 | 5.0 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 1.4 | 17.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 1.3 | 4.0 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 1.2 | 6.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 1.1 | 6.9 | GO:0030421 | defecation(GO:0030421) |
| 0.8 | 8.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.8 | 3.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.8 | 9.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.8 | 7.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.8 | 3.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.7 | 2.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.7 | 4.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.7 | 2.2 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.6 | 2.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.6 | 7.9 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.6 | 2.9 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.6 | 6.4 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.6 | 2.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 2.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.5 | 8.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.5 | 4.5 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.5 | 3.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.5 | 1.5 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.5 | 1.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.5 | 4.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.5 | 1.4 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.4 | 3.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.4 | 1.7 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.4 | 6.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 2.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.4 | 3.9 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.4 | 1.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.4 | 1.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.4 | 2.2 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.4 | 5.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 1.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 | 2.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.3 | 3.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 2.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 6.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 1.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 3.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 0.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 2.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 4.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 1.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.3 | 4.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.3 | 2.2 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 1.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 1.3 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.3 | 5.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 2.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 4.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 24.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.2 | 1.4 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 1.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 1.6 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 6.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 1.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 5.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 2.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 3.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 0.8 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 5.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 4.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 5.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 4.9 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 2.0 | GO:0006189 | brainstem development(GO:0003360) 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 1.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 4.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 13.2 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 3.0 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 1.3 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 2.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 5.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 6.5 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 14.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.9 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 1.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 5.4 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 1.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 2.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 3.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 3.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 2.0 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 1.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.3 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 1.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 6.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 2.9 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 6.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.6 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.4 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.6 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.6 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 6.3 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 11.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.0 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 3.3 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) |
| 0.0 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 3.6 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 5.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 1.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 7.3 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.2 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 1.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 2.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 1.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.7 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 4.9 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.9 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 1.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 7.3 | GO:0010608 | posttranscriptional regulation of gene expression(GO:0010608) |
| 0.0 | 4.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 23.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 2.2 | 24.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 1.6 | 13.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.4 | 17.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.3 | 5.0 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 1.0 | 7.8 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.9 | 9.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.9 | 3.7 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.6 | 2.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.5 | 2.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.5 | 6.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.5 | 9.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 1.9 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.4 | 6.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 5.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 1.8 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.4 | 2.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 2.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 2.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.3 | 2.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 2.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 7.4 | GO:0030054 | cell junction(GO:0030054) |
| 0.2 | 2.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 2.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.2 | 0.8 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 3.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 7.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 1.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 16.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 3.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 3.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 2.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 7.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 8.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 4.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 2.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 4.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 5.8 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 17.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 1.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 2.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 2.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 6.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 5.2 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 3.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 3.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.3 | 4.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 1.3 | 3.9 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 1.2 | 8.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.0 | 5.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.0 | 2.9 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 1.0 | 5.9 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.8 | 6.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.8 | 3.8 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.7 | 3.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.7 | 2.2 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.7 | 17.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.7 | 8.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.6 | 2.3 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.5 | 4.8 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.5 | 2.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.5 | 2.5 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.5 | 2.0 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.5 | 3.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.5 | 3.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.5 | 1.4 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.4 | 5.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 7.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 1.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.3 | 4.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.3 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.3 | 2.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 2.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 8.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 0.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 2.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.3 | 2.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 5.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 2.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 6.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 5.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 1.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 2.8 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 2.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 1.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 3.7 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 1.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 10.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 15.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.6 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.2 | 2.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 0.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 2.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 1.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 5.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.4 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 1.0 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 3.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 4.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.4 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 1.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.4 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.1 | 2.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 31.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 3.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 2.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 5.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 3.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 3.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 3.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 4.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 2.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 8.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 4.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 6.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 7.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 3.3 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 9.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 5.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 9.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 17.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 10.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 3.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 18.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 8.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 11.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 7.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 15.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 4.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 6.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 3.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 8.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 8.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 8.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 8.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.9 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 22.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 28.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.4 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 3.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 4.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 6.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 1.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 6.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 6.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 5.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 5.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 8.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 4.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 5.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 4.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 4.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 5.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 4.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |