Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
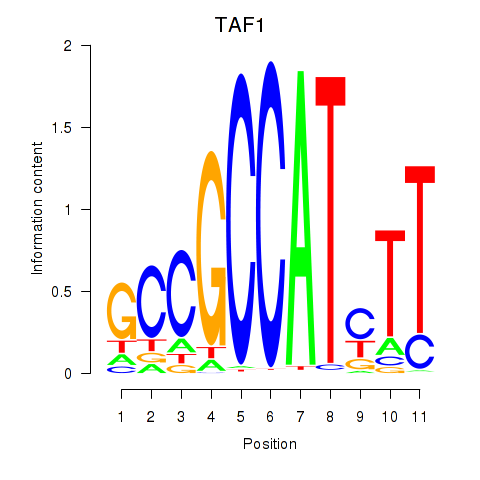
Results for TAF1
Z-value: 5.36
Transcription factors associated with TAF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TAF1
|
ENSG00000147133.11 | TATA-box binding protein associated factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TAF1 | hg19_v2_chrX_+_70586082_70586114 | 0.97 | 1.0e-12 | Click! |
Activity profile of TAF1 motif
Sorted Z-values of TAF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TAF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 2.5 | 10.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 2.3 | 7.0 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 2.3 | 29.9 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 2.2 | 8.9 | GO:0019046 | release from viral latency(GO:0019046) |
| 2.1 | 6.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 2.1 | 8.5 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 2.0 | 5.9 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 1.8 | 1.8 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 1.8 | 7.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.7 | 5.2 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 1.7 | 8.3 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 1.6 | 3.2 | GO:0034091 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 1.6 | 8.0 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.5 | 4.6 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 1.5 | 4.4 | GO:0044209 | AMP salvage(GO:0044209) |
| 1.5 | 7.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.5 | 4.4 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 1.4 | 4.3 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 1.4 | 8.6 | GO:0070541 | response to platinum ion(GO:0070541) |
| 1.4 | 5.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.4 | 12.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 1.4 | 4.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 1.3 | 5.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.3 | 5.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.3 | 6.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 1.3 | 7.6 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 1.2 | 3.5 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 1.2 | 4.6 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 1.2 | 4.6 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 1.1 | 1.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 1.1 | 7.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.1 | 11.0 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 1.1 | 3.3 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 1.1 | 15.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.1 | 4.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 1.0 | 10.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 1.0 | 7.2 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 1.0 | 4.1 | GO:1904501 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 1.0 | 1.0 | GO:0010632 | regulation of epithelial cell migration(GO:0010632) |
| 1.0 | 5.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.0 | 7.0 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 1.0 | 4.9 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 1.0 | 6.7 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.9 | 5.5 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.9 | 5.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.8 | 8.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.8 | 11.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.8 | 7.5 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.8 | 4.9 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.8 | 9.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.8 | 2.5 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.8 | 4.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.8 | 4.0 | GO:0048319 | mesoderm migration involved in gastrulation(GO:0007509) axial mesoderm morphogenesis(GO:0048319) |
| 0.8 | 11.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.8 | 10.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.8 | 6.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.8 | 2.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.8 | 3.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.8 | 9.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.8 | 3.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.7 | 7.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.7 | 2.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.7 | 5.8 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.7 | 2.9 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.7 | 2.8 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.7 | 6.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.7 | 0.7 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.7 | 4.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.7 | 2.8 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.7 | 4.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.7 | 4.8 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.7 | 2.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.7 | 3.9 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.7 | 2.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.7 | 9.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.6 | 2.6 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.6 | 14.6 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.6 | 1.9 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.6 | 5.6 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.6 | 4.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.6 | 3.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.6 | 29.8 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) |
| 0.6 | 11.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 1.8 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.6 | 2.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.6 | 7.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.6 | 4.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.6 | 2.9 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.6 | 5.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.6 | 1.7 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.6 | 6.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.6 | 4.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.6 | 2.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.6 | 2.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.6 | 3.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.6 | 0.6 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.5 | 1.6 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.5 | 1.6 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.5 | 2.6 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.5 | 2.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.5 | 5.7 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.5 | 3.6 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.5 | 9.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.5 | 2.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.5 | 5.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 0.5 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.5 | 4.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.5 | 4.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.5 | 4.9 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.5 | 1.5 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.5 | 3.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 3.9 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.5 | 1.5 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.5 | 8.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 1.9 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.5 | 4.8 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.5 | 1.9 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.5 | 2.9 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.5 | 1.4 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.5 | 3.8 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 | 5.7 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.5 | 1.4 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.5 | 4.7 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.5 | 3.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 2.3 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.5 | 2.3 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.5 | 1.4 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.5 | 4.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.5 | 1.4 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.5 | 1.4 | GO:0048007 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 3.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.4 | 5.8 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.4 | 0.9 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.4 | 19.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.4 | 1.3 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.4 | 8.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.4 | 1.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.4 | 9.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.4 | 1.7 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.4 | 1.7 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.4 | 0.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.4 | 1.7 | GO:0043622 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.4 | 1.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 1.2 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.4 | 1.2 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.4 | 2.8 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.4 | 4.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.4 | 2.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.4 | 5.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.4 | 2.8 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.4 | 4.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.4 | 8.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.4 | 10.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.4 | 5.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.4 | 1.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.4 | 3.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.4 | 2.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.4 | 1.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.4 | 5.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 7.5 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.4 | 0.7 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.3 | 0.3 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.3 | 5.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.3 | 1.7 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.3 | 3.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 2.0 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 1.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.3 | 1.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.3 | 2.6 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.3 | 2.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.3 | 9.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.3 | 1.9 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 0.6 | GO:0043201 | response to leucine(GO:0043201) |
| 0.3 | 1.9 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 1.9 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 6.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 0.6 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 6.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.3 | 3.7 | GO:2000507 | cap-dependent translational initiation(GO:0002191) positive regulation of energy homeostasis(GO:2000507) |
| 0.3 | 2.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 1.5 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.3 | 4.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.3 | 1.2 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 2.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.3 | 1.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.3 | 1.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.3 | 1.2 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 | 1.5 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 8.3 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.3 | 2.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 2.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 1.8 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.3 | 1.7 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.3 | 2.0 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.3 | 1.4 | GO:0090169 | regulation of spindle assembly(GO:0090169) |
| 0.3 | 4.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 0.9 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.3 | 1.4 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.3 | 3.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 3.1 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.3 | 24.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.3 | 3.8 | GO:0036124 | histone H3-K9 trimethylation(GO:0036124) |
| 0.3 | 1.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.3 | 2.4 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.3 | 1.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.3 | 1.8 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.3 | 0.8 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 4.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 1.5 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.3 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 1.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 1.2 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 1.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 3.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 6.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 2.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 1.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 3.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 3.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 2.9 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 3.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 2.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.2 | 0.9 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 0.9 | GO:0051572 | negative regulation of histone H3-K36 methylation(GO:0000415) negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.2 | 6.8 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 0.5 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.2 | 4.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.2 | 0.7 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 5.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 1.4 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.2 | 2.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 2.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 1.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 2.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 2.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 3.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.8 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 8.7 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 1.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 1.1 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.2 | 1.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 2.8 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 1.0 | GO:0006167 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) AMP biosynthetic process(GO:0006167) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.4 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.2 | 1.9 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.2 | 0.6 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.2 | 5.9 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.2 | 5.7 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) |
| 0.2 | 1.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 1.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 7.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 1.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 2.5 | GO:0016559 | peroxisome fission(GO:0016559) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 1.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 1.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 0.6 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.2 | 5.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 1.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 2.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 3.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 2.0 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 4.8 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.2 | 2.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 3.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 12.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 3.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 10.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 0.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 0.7 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.2 | 0.7 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.2 | 2.8 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.2 | 0.5 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.2 | 0.5 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.2 | 2.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 1.5 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 0.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 3.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 1.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.5 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.2 | 1.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 0.9 | GO:0061087 | regulation of histone H3-K27 methylation(GO:0061085) positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 2.8 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.2 | 5.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 3.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 1.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 5.0 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.2 | 1.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 6.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.2 | 0.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 4.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.9 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 1.8 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 4.8 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 2.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.6 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 1.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 2.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.6 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.7 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.6 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 1.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 4.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.7 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 1.4 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 0.7 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.1 | 4.7 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.7 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.3 | GO:0071317 | cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.1 | 0.5 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 3.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.7 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) DNA methylation or demethylation(GO:0044728) |
| 0.1 | 1.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.4 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.6 | GO:2000672 | response to sorbitol(GO:0072708) cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.6 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 2.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.1 | 1.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.6 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 1.5 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.5 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 0.6 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 2.8 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.6 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 1.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 3.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 1.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.7 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 7.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.0 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 2.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 2.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 9.7 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.5 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 1.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 6.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.6 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.1 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.3 | GO:0031577 | spindle checkpoint(GO:0031577) |
| 0.1 | 2.0 | GO:1901990 | regulation of mitotic cell cycle phase transition(GO:1901990) |
| 0.1 | 0.5 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 8.2 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.7 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 2.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 2.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 1.2 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 1.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 3.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 5.3 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 2.0 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 1.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 1.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 5.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.2 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 2.4 | GO:0043928 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 4.8 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.1 | 0.4 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 6.6 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.8 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 3.3 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 0.3 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 1.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.5 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 3.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 6.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 1.0 | GO:1902745 | positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 2.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 0.7 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 2.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.6 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 2.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.5 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 0.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.4 | GO:0043473 | pigmentation(GO:0043473) |
| 0.1 | 9.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.6 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.1 | 0.3 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.1 | 1.3 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 0.6 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.1 | 5.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.7 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.1 | 1.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.7 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 1.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.9 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 1.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.1 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.1 | 1.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.1 | 0.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 6.2 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 1.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 1.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 1.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.7 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 2.1 | GO:0090659 | walking behavior(GO:0090659) |
| 0.1 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 2.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.8 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 3.9 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 4.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 2.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 3.3 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.9 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 1.2 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 4.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 4.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 2.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.8 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 3.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.7 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:1903376 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) |
| 0.0 | 1.1 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 2.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.6 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 5.6 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 3.1 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 0.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.6 | GO:0030888 | regulation of B cell proliferation(GO:0030888) |
| 0.0 | 2.3 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.6 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.2 | GO:2000142 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.4 | GO:0051297 | centrosome cycle(GO:0007098) centrosome organization(GO:0051297) |
| 0.0 | 0.8 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 1.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.2 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 1.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.8 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.2 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) |
| 0.0 | 0.5 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.3 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 1.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.7 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.5 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.7 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.0 | 3.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:1900077 | negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.4 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.3 | GO:1901185 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 7.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.7 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 2.4 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.8 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.0 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.6 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 5.5 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.9 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.9 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 24.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 2.9 | 28.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 2.3 | 7.0 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 2.1 | 8.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 2.0 | 40.5 | GO:0097227 | sperm annulus(GO:0097227) |
| 2.0 | 10.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.7 | 5.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 1.4 | 1.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 1.4 | 23.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 1.3 | 5.2 | GO:0031213 | RSF complex(GO:0031213) |
| 1.2 | 3.7 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.2 | 4.9 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 1.2 | 8.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.1 | 30.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.1 | 5.6 | GO:0071942 | XPC complex(GO:0071942) |
| 1.1 | 6.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.0 | 4.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.0 | 5.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.0 | 4.0 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.0 | 10.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.9 | 2.7 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.9 | 3.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.8 | 12.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.8 | 2.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.8 | 9.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.8 | 3.0 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.7 | 2.2 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.7 | 1.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.7 | 2.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.7 | 10.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.7 | 4.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.7 | 2.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.7 | 7.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 1.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.6 | 4.3 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.6 | 1.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 15.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.6 | 3.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.6 | 16.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.6 | 3.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.6 | 4.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.6 | 1.7 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.5 | 5.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 1.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.5 | 3.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.5 | 13.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 11.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.5 | 3.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.5 | 1.5 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.5 | 4.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 7.9 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.5 | 2.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.5 | 7.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 3.7 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.4 | 2.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 3.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.4 | 1.7 | GO:1990745 | EARP complex(GO:1990745) |
| 0.4 | 1.2 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.4 | 5.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 5.7 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.4 | 2.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.4 | 7.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 3.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.4 | 2.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 4.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.4 | 2.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 1.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.4 | 1.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.4 | 1.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.4 | 3.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 1.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.3 | 4.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 1.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 3.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.3 | 7.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 1.6 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.3 | 1.3 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.3 | 6.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 1.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 4.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 3.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.3 | 5.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 5.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 2.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 2.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 1.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 1.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 0.9 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.3 | 3.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 5.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 14.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 1.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 2.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 8.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 2.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 3.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 5.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.2 | 3.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 0.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 7.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 2.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.7 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.2 | 0.7 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 2.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 13.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 3.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 7.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 0.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 3.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 24.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 2.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.8 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 7.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 2.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.4 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.2 | 6.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 1.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 6.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 3.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 1.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.2 | 2.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 2.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 9.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 1.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 1.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 1.5 | GO:0090576 | transcription factor TFIIIC complex(GO:0000127) RNA polymerase III transcription factor complex(GO:0090576) |
| 0.2 | 1.0 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 1.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 18.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 2.5 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 1.1 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 2.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 6.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 11.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 5.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 28.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 11.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 9.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 3.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.8 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 2.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 5.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.9 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.1 | 5.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 3.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 3.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 7.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 5.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 9.6 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 10.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 6.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 2.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 13.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 3.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 3.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 24.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 3.1 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 2.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 3.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 9.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 7.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 19.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.7 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 19.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 8.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.8 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 5.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 5.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 6.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 4.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.9 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 5.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 3.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 5.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 7.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 7.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 3.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.6 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.1 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 6.6 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 0.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 3.9 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 2.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 3.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.2 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 2.3 | 9.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 2.3 | 11.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 2.2 | 8.6 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 2.0 | 8.0 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 1.9 | 9.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.8 | 5.3 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 1.7 | 5.2 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 1.7 | 5.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 1.7 | 5.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 1.7 | 6.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.3 | 5.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 1.2 | 4.9 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 1.1 | 8.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.1 | 4.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 1.1 | 6.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 1.0 | 3.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 1.0 | 8.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.0 | 3.9 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 1.0 | 2.9 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 1.0 | 3.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.0 | 5.8 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.9 | 0.9 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.9 | 11.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.9 | 2.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.8 | 3.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.8 | 7.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.8 | 2.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.8 | 8.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.8 | 3.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.7 | 5.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.7 | 15.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.7 | 2.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.7 | 4.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.7 | 3.6 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.7 | 3.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.7 | 2.8 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.7 | 6.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.6 | 10.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.6 | 1.9 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.6 | 5.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.6 | 4.4 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.6 | 1.8 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.6 | 1.8 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.6 | 4.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.6 | 2.3 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.6 | 7.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.6 | 4.6 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 3.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.6 | 2.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.6 | 1.7 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.6 | 3.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.6 | 16.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.5 | 2.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.5 | 3.8 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.5 | 2.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.5 | 1.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.5 | 2.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 2.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.5 | 5.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.5 | 1.5 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.5 | 4.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.5 | 2.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.5 | 1.4 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.5 | 6.0 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.5 | 39.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.5 | 3.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.4 | 4.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 3.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 1.3 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.4 | 3.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.4 | 22.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.4 | 4.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 2.9 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.4 | 1.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.4 | 0.8 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.4 | 1.2 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.4 | 5.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 3.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.4 | 8.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 1.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.4 | 11.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.4 | 1.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.4 | 2.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 11.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 6.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.3 | 1.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.3 | 4.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 1.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.3 | 1.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 9.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 2.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 5.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 8.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 15.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 1.0 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.3 | 8.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 2.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.3 | 2.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 29.5 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 2.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 3.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 2.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.3 | 12.9 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.3 | 2.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 13.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 2.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 1.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 1.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.3 | 3.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 26.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 1.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 6.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 2.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.3 | 5.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 1.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 2.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.3 | 4.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 5.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 3.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 1.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 7.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 10.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 2.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 1.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 1.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 10.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 2.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 0.5 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.2 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 2.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.2 | 12.4 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 5.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 2.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 3.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 1.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 0.6 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.2 | 1.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 1.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 9.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 12.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 2.3 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.2 | 2.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 6.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 0.6 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.2 | 16.2 | GO:0034212 | peptide N-acetyltransferase activity(GO:0034212) |
| 0.2 | 2.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 2.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 3.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.2 | 6.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 4.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 8.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 23.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 0.7 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 9.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.2 | 6.5 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.2 | 2.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 0.5 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 22.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 3.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 21.4 | GO:0101005 | ubiquitinyl hydrolase activity(GO:0101005) |
| 0.2 | 0.9 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 2.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 1.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 6.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 5.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 3.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 4.4 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.1 | 2.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 4.0 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 3.7 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 1.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 3.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.5 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 6.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 5.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 7.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.8 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 3.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 3.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 12.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 9.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 5.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 1.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 5.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 2.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 8.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.1 | 1.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 17.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 45.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 2.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 27.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 1.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 3.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 2.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 4.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.7 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.6 | GO:0003916 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 5.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 3.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.6 | GO:0000988 | transcription factor activity, protein binding(GO:0000988) transcription factor activity, transcription factor binding(GO:0000989) |
| 0.1 | 3.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) SAM domain binding(GO:0032093) |
| 0.1 | 0.5 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 1.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.6 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 5.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.9 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 2.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 22.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 13.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 5.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 2.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0052811 | 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) |
| 0.0 | 0.9 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 5.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0000829 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 4.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 2.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.3 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 23.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.5 | 15.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.5 | 17.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 38.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 24.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 47.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.3 | 6.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 20.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 28.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 13.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 9.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 8.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 4.7 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 16.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 8.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 6.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 8.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 7.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 13.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 12.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 10.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 6.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 4.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 3.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 5.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 7.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 5.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 6.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 9.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 7.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 3.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 6.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 4.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 3.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 2.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 5.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.5 | 7.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.5 | 4.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.5 | 15.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.5 | 11.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 15.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 9.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 47.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.4 | 9.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 8.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 31.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.4 | 11.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.4 | 4.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.4 | 11.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 17.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 3.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 1.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 7.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.3 | 7.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 12.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 2.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 18.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 3.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.3 | 1.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 1.5 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.3 | 10.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 6.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 5.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 11.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 8.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 7.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 12.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 8.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 18.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 1.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 6.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 2.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 26.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 22.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 3.7 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.2 | 10.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.2 | 11.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 2.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 6.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 10.8 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.2 | 8.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 5.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 12.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 5.5 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.2 | 2.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 3.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 4.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 5.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 4.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 4.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 3.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 14.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 4.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 2.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 2.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 5.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 2.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 5.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 16.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 3.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 18.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 1.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 2.6 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |