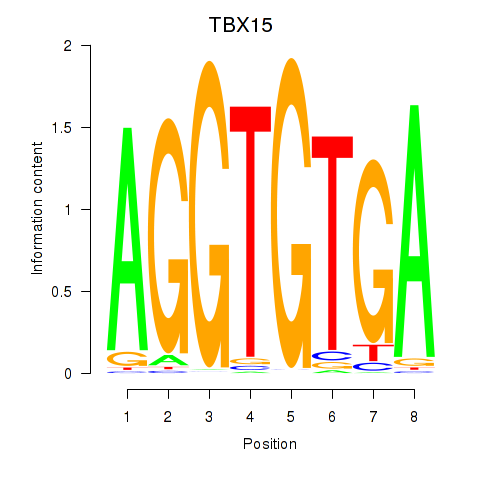
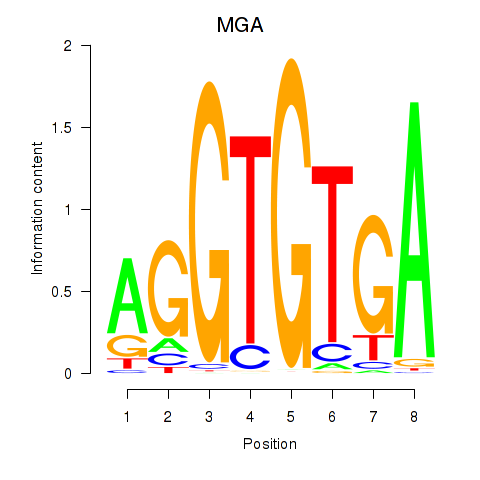
| 0.5 |
4.7 |
GO:1904379 |
protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.4 |
1.8 |
GO:1905205 |
positive regulation of connective tissue replacement(GO:1905205) |
| 0.4 |
2.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 |
1.0 |
GO:0070563 |
negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 |
0.8 |
GO:0035627 |
ceramide transport(GO:0035627) |
| 0.2 |
0.6 |
GO:0061433 |
cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.2 |
0.7 |
GO:0010983 |
positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 |
0.7 |
GO:0006781 |
succinyl-CoA pathway(GO:0006781) |
| 0.2 |
0.9 |
GO:0060574 |
intestinal epithelial cell maturation(GO:0060574) |
| 0.1 |
0.7 |
GO:1904504 |
regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.1 |
0.9 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 |
1.3 |
GO:1903764 |
regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 |
0.5 |
GO:2001160 |
regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 |
0.4 |
GO:1903570 |
regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 |
0.7 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
0.6 |
GO:0030807 |
positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 |
0.4 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.7 |
GO:0035879 |
plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.4 |
GO:1903691 |
positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 |
1.3 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
1.0 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.1 |
0.3 |
GO:0018315 |
molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 |
0.3 |
GO:2000681 |
negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 |
0.6 |
GO:1902962 |
regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 |
0.5 |
GO:2000670 |
positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 |
0.8 |
GO:1901315 |
negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 |
0.5 |
GO:0039019 |
pronephric nephron development(GO:0039019) |
| 0.1 |
0.3 |
GO:0046379 |
extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 |
0.3 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.1 |
0.2 |
GO:0006117 |
acetaldehyde metabolic process(GO:0006117) |
| 0.1 |
0.2 |
GO:0021589 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.4 |
GO:1900224 |
positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 |
0.3 |
GO:1904501 |
glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 |
0.5 |
GO:2001166 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.5 |
GO:1903575 |
cornified envelope assembly(GO:1903575) |
| 0.1 |
0.5 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.3 |
GO:0021623 |
optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 |
0.3 |
GO:0098582 |
innate vocalization behavior(GO:0098582) |
| 0.1 |
0.2 |
GO:0061536 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 |
0.4 |
GO:0030421 |
defecation(GO:0030421) |
| 0.1 |
0.9 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.5 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.3 |
GO:0010760 |
negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 |
0.7 |
GO:1990416 |
cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.5 |
GO:1905232 |
cellular response to L-glutamate(GO:1905232) |
| 0.1 |
0.2 |
GO:0035522 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.2 |
GO:0031247 |
actin rod assembly(GO:0031247) |
| 0.1 |
0.7 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.3 |
GO:0003069 |
age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 |
0.3 |
GO:0019470 |
4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 |
0.3 |
GO:1903588 |
negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 |
0.2 |
GO:0002904 |
positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 |
0.2 |
GO:0097010 |
eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 |
0.3 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.2 |
GO:0021603 |
cranial nerve formation(GO:0021603) |
| 0.0 |
0.3 |
GO:0051414 |
response to cortisol(GO:0051414) |
| 0.0 |
0.1 |
GO:1901340 |
negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 |
0.8 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.3 |
GO:0038060 |
nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 |
0.5 |
GO:0035524 |
proline transmembrane transport(GO:0035524) |
| 0.0 |
0.3 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 |
0.1 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.0 |
0.2 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 |
0.1 |
GO:0060532 |
bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 |
0.3 |
GO:2000490 |
negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 |
0.2 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.4 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
0.2 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.2 |
GO:0044855 |
plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 |
0.6 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.2 |
GO:1902766 |
skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 |
0.1 |
GO:0021827 |
cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 |
0.2 |
GO:0097475 |
motor neuron migration(GO:0097475) |
| 0.0 |
0.1 |
GO:1903824 |
regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 |
0.1 |
GO:0040040 |
thermosensory behavior(GO:0040040) |
| 0.0 |
0.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 |
0.1 |
GO:0033861 |
negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 |
0.4 |
GO:1904352 |
positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 |
0.1 |
GO:1903045 |
neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 |
0.1 |
GO:0044210 |
'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.8 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.6 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.5 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.0 |
0.6 |
GO:0035360 |
positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.1 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.0 |
0.2 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.1 |
GO:0048294 |
negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0035290 |
trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 |
0.1 |
GO:0000915 |
assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 |
1.2 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.0 |
0.3 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.1 |
GO:0021834 |
chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 |
0.1 |
GO:1902595 |
regulation of DNA replication origin binding(GO:1902595) |
| 0.0 |
0.5 |
GO:0031629 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.2 |
GO:1903593 |
regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 |
0.7 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 |
0.4 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.0 |
0.3 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.2 |
GO:0099590 |
neurotransmitter receptor internalization(GO:0099590) |
| 0.0 |
1.6 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.1 |
GO:1904721 |
negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 |
0.1 |
GO:0060067 |
chemoattraction of serotonergic neuron axon(GO:0036517) cervix development(GO:0060067) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 |
0.2 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
0.1 |
GO:1990737 |
response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 |
0.1 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.2 |
GO:0001915 |
negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 |
0.2 |
GO:0098814 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 |
0.1 |
GO:0051461 |
positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.0 |
GO:1990641 |
response to iron ion starvation(GO:1990641) |
| 0.0 |
0.5 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.1 |
GO:0008204 |
ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 |
0.1 |
GO:2000317 |
negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 |
0.2 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) |
| 0.0 |
0.6 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.1 |
GO:0072752 |
cellular response to rapamycin(GO:0072752) |
| 0.0 |
0.3 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.1 |
GO:0006617 |
SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 |
0.1 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.0 |
0.1 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.0 |
0.4 |
GO:2000059 |
negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 |
0.4 |
GO:0060211 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:0051121 |
hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 |
0.1 |
GO:0010645 |
regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 |
0.2 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 |
1.4 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.1 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 |
0.4 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.2 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.1 |
GO:1904636 |
response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 |
0.2 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.0 |
0.1 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 |
0.3 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.2 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.2 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.1 |
GO:0003366 |
cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 |
0.1 |
GO:0097101 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 |
0.1 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 |
0.2 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.0 |
GO:0090678 |
metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 |
2.1 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.1 |
GO:0051096 |
positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.1 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.1 |
GO:0048341 |
paraxial mesoderm formation(GO:0048341) |
| 0.0 |
0.1 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0090344 |
negative regulation of cell aging(GO:0090344) |
| 0.0 |
0.1 |
GO:0055118 |
intracellular sequestering of iron ion(GO:0006880) negative regulation of cardiac muscle contraction(GO:0055118) sequestering of iron ion(GO:0097577) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.1 |
GO:2001288 |
positive regulation of caveolin-mediated endocytosis(GO:2001288) |