Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
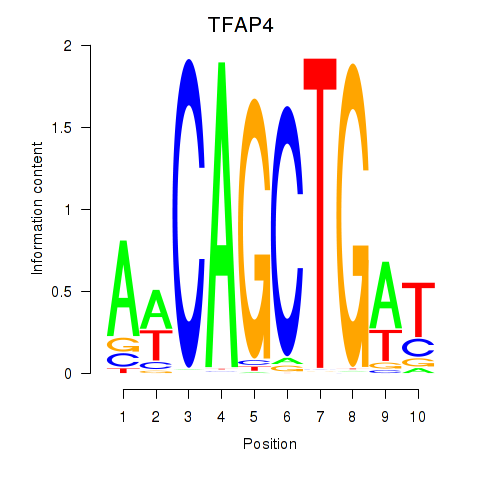
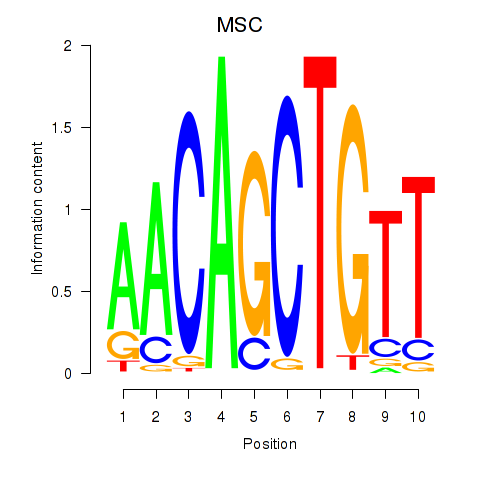
Results for TFAP4_MSC
Z-value: 2.75
Transcription factors associated with TFAP4_MSC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP4
|
ENSG00000090447.7 | transcription factor AP-4 |
|
MSC
|
ENSG00000178860.8 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSC | hg19_v2_chr8_-_72756667_72756736 | -0.89 | 2.1e-07 | Click! |
| TFAP4 | hg19_v2_chr16_-_4323015_4323076 | -0.86 | 1.1e-06 | Click! |
Activity profile of TFAP4_MSC motif
Sorted Z-values of TFAP4_MSC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP4_MSC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 2.2 | 13.0 | GO:0018032 | protein amidation(GO:0018032) |
| 1.7 | 5.0 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 1.3 | 4.0 | GO:0048627 | myoblast development(GO:0048627) |
| 1.3 | 5.2 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 1.2 | 3.6 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 1.1 | 4.5 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 1.0 | 5.2 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 1.0 | 3.0 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.9 | 3.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.8 | 9.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.8 | 9.8 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.8 | 2.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.7 | 2.2 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.7 | 3.5 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.7 | 2.1 | GO:0048003 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.7 | 2.7 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.6 | 12.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.6 | 1.8 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.6 | 1.7 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.5 | 5.5 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.5 | 2.7 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.5 | 1.6 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.5 | 1.6 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.5 | 11.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 2.6 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.5 | 2.0 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.5 | 4.9 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.5 | 2.9 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.5 | 1.9 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.4 | 2.2 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.4 | 2.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 2.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.4 | 4.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 2.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 5.0 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.4 | 11.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.4 | 1.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 1.9 | GO:0071874 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.4 | 1.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 2.2 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.4 | 1.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.3 | 1.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 1.6 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.3 | 4.4 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.3 | 4.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 1.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.3 | 1.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.3 | 2.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 1.2 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.2 | 1.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 1.2 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.2 | 0.7 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.2 | 0.7 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.2 | 2.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.9 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.2 | 2.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 2.4 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.2 | 1.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 0.8 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.2 | 5.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 3.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 3.9 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 1.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.2 | 1.7 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.8 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.7 | GO:0072254 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.2 | 0.9 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 3.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 1.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 1.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.0 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 0.5 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 0.7 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 0.2 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 1.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.8 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 1.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 2.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.9 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.5 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.4 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 2.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 3.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.9 | GO:0097319 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 1.7 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.4 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 3.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.6 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 1.4 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 5.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 1.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.6 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 2.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.4 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.7 | GO:0071971 | extracellular exosome assembly(GO:0071971) regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 0.5 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 0.4 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 1.2 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 3.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 2.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 5.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 3.9 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 1.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 2.8 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 2.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 1.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.8 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 1.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.5 | GO:0061086 | regulation of mRNA export from nucleus(GO:0010793) negative regulation of histone H3-K27 methylation(GO:0061086) chromatin maintenance(GO:0070827) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 1.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 0.6 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.1 | 2.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.2 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 3.2 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.6 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.3 | GO:0018879 | insecticide metabolic process(GO:0017143) biphenyl metabolic process(GO:0018879) phthalate metabolic process(GO:0018963) response to luteinizing hormone(GO:0034699) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.1 | 0.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 2.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 2.6 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 2.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 1.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 2.2 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 4.0 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 1.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.4 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.1 | 1.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 4.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 4.5 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.0 | 0.2 | GO:0014870 | response to muscle inactivity(GO:0014870) |
| 0.0 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 1.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 1.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 5.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.5 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 1.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 2.2 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 3.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 1.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 4.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.1 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 2.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.5 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 1.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 1.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 1.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.6 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.7 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.3 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 1.9 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 3.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.1 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 4.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 2.9 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.0 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 4.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.7 | GO:0016032 | viral process(GO:0016032) multi-organism cellular process(GO:0044764) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.8 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 2.9 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 1.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 1.4 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.3 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.1 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.0 | 0.8 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 2.0 | GO:0031424 | keratinization(GO:0031424) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.0 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.7 | 2.0 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.5 | 2.5 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.5 | 2.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.5 | 7.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.4 | 3.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.4 | 3.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 1.0 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 5.5 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 2.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 2.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 2.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 2.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 4.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 2.9 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 0.9 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 5.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 3.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 1.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 1.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 2.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 0.8 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.2 | 3.0 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 2.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 2.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 6.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.7 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 14.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 3.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.5 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 2.6 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 21.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 5.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 8.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 6.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 11.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.7 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 24.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 4.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 7.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.8 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 4.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 1.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 2.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 3.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 8.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 21.8 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 3.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 4.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 15.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.0 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.2 | 9.9 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 1.2 | 4.7 | GO:0008431 | vitamin E binding(GO:0008431) |
| 1.0 | 5.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.0 | 3.0 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.9 | 3.8 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.9 | 3.5 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.9 | 2.6 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.8 | 12.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.7 | 3.9 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.6 | 2.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.6 | 1.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.5 | 2.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.4 | 2.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 5.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 2.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 2.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 1.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.3 | 9.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 0.9 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.3 | 1.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 2.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 5.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 1.7 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.8 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.2 | 3.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 15.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 1.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 7.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 5.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 1.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 0.5 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 9.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 6.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 2.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 4.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 5.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 2.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 16.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 3.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.4 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.9 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.5 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 4.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 5.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 2.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 5.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 3.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 2.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 3.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 14.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 3.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 2.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 2.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 5.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 1.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 2.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 7.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 4.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 3.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 6.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 4.1 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.2 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 3.6 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.0 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 7.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 6.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 5.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 2.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.3 | GO:0046332 | SMAD binding(GO:0046332) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 21.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 6.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 12.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 11.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 5.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 5.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 9.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 5.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 1.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 4.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 4.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 5.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 11.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 5.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 5.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 6.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 2.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 9.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 13.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 4.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 11.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.5 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 2.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.9 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 3.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 2.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |