Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
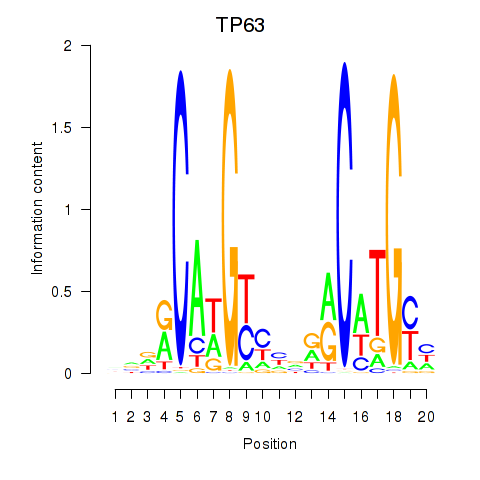
Results for TP63
Z-value: 6.51
Transcription factors associated with TP63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP63
|
ENSG00000073282.8 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP63 | hg19_v2_chr3_+_189507460_189507471 | 0.99 | 3.0e-19 | Click! |
Activity profile of TP63 motif
Sorted Z-values of TP63 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TP63
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 84.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 8.5 | 33.9 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 8.3 | 33.0 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 8.1 | 32.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 5.4 | 16.3 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 4.9 | 14.7 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 4.6 | 13.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 4.5 | 18.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 3.3 | 26.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.7 | 8.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 2.7 | 13.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 2.2 | 9.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.6 | 6.5 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 1.6 | 4.8 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 1.6 | 9.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.6 | 37.9 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 1.4 | 10.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.4 | 7.0 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 1.3 | 6.6 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.3 | 32.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 1.2 | 7.4 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 1.2 | 6.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 1.2 | 4.8 | GO:0045401 | response to molecule of fungal origin(GO:0002238) serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 1.2 | 7.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.1 | 10.2 | GO:0003383 | apical constriction(GO:0003383) |
| 1.1 | 4.4 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 1.1 | 27.6 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 1.1 | 72.7 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 1.0 | 5.2 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 1.0 | 4.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 1.0 | 5.0 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.0 | 6.9 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 1.0 | 46.3 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 1.0 | 4.8 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.8 | 8.7 | GO:0090032 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.8 | 3.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.8 | 7.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.7 | 2.2 | GO:0071262 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.7 | 8.0 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.7 | 2.2 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.7 | 5.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.7 | 14.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.6 | 1.9 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.6 | 8.0 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.6 | 2.4 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.6 | 16.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 2.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 1.5 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.5 | 8.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.5 | 2.4 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 | 13.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.4 | 4.6 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.4 | 1.2 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve formation(GO:0021650) |
| 0.4 | 3.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.4 | 1.6 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.4 | 7.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.4 | 2.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.4 | 13.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 2.5 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.3 | 8.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 11.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 1.4 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 5.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.3 | 7.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 9.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.3 | 24.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 10.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 1.2 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.3 | 4.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 3.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.3 | 1.9 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 3.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.2 | 4.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.6 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 1.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 | 1.3 | GO:0031860 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 2.3 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 5.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 3.3 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.2 | 4.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 2.3 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 0.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 2.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 10.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 2.9 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 3.9 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.2 | 1.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.2 | 0.5 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 1.1 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 3.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 6.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 23.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 3.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.4 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 7.4 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 3.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.4 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 3.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 9.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 14.2 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 4.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 1.3 | GO:1903943 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 4.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 3.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 2.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.2 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 | 1.3 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.5 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 1.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 2.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 3.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 6.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 4.5 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.1 | 4.2 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 2.4 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 1.6 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 5.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 2.2 | GO:0016569 | covalent chromatin modification(GO:0016569) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 2.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 1.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 9.2 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 3.5 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 1.4 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.8 | GO:0070271 | protein complex assembly(GO:0006461) protein complex biogenesis(GO:0070271) |
| 0.0 | 1.9 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 2.7 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 1.7 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 3.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 2.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.3 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 2.1 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 1.9 | 7.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.7 | 5.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 1.6 | 8.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 1.3 | 9.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.3 | 13.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.1 | 3.3 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 1.0 | 34.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 1.0 | 4.8 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.9 | 3.8 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.8 | 2.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.7 | 4.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 54.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 13.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.5 | 6.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.5 | 8.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 7.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 5.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.5 | 4.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.4 | 10.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 23.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 3.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.3 | 8.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 1.6 | GO:0071942 | XPC complex(GO:0071942) |
| 0.3 | 11.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.3 | 3.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 21.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 6.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.3 | 1.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 4.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 16.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 0.9 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 11.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 17.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 14.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 4.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 4.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 4.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.7 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.1 | 3.0 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 6.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 7.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 61.6 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 8.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 4.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 3.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 26.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 4.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.9 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 1.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 8.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 6.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.4 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 3.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 109.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 5.4 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 6.8 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 84.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 5.5 | 33.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 5.4 | 16.3 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 4.3 | 12.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 3.3 | 10.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 2.2 | 9.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 2.0 | 8.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 2.0 | 32.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 1.9 | 11.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 1.5 | 26.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 1.3 | 14.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.3 | 6.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.2 | 4.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.2 | 7.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.1 | 15.3 | GO:0089720 | caspase binding(GO:0089720) |
| 1.0 | 4.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 1.0 | 5.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.0 | 4.8 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.9 | 2.8 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.8 | 7.5 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.8 | 5.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.8 | 14.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.7 | 23.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.7 | 4.6 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.6 | 34.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 2.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 7.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.6 | 4.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.6 | 82.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 1.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.5 | 1.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.5 | 1.6 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.5 | 27.6 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.5 | 5.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.5 | 1.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.5 | 7.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 8.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 8.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.4 | 10.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 2.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 8.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 2.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 11.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 2.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.5 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 6.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 3.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 7.2 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.3 | 1.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 5.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 61.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 8.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 5.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 2.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 11.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 48.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 2.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 24.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 5.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 3.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 9.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 21.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 3.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 48.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 2.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 5.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 4.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 3.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 3.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.9 | GO:0008301 | four-way junction DNA binding(GO:0000400) DNA binding, bending(GO:0008301) |
| 0.1 | 1.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 3.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 3.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 4.2 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 2.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 3.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 22.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 2.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 1.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 8.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 11.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 19.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.0 | GO:0008266 | poly-pyrimidine tract binding(GO:0008187) poly(U) RNA binding(GO:0008266) |
| 0.0 | 4.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.5 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 103.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.8 | 34.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.8 | 16.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.6 | 13.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 55.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.5 | 18.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 8.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 11.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 8.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 17.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 7.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 8.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 14.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 3.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 6.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 4.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 20.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 4.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 11.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 39.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.1 | 18.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 16.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.6 | 13.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.5 | 14.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 9.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 86.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.3 | 7.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 6.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.3 | 26.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 8.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 8.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 5.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 7.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 4.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 10.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 4.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 4.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 4.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 9.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 4.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 14.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 6.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 4.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |