Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for TP73
Z-value: 2.01
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.10 | tumor protein p73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP73 | hg19_v2_chr1_+_3598871_3598930 | 0.23 | 3.3e-01 | Click! |
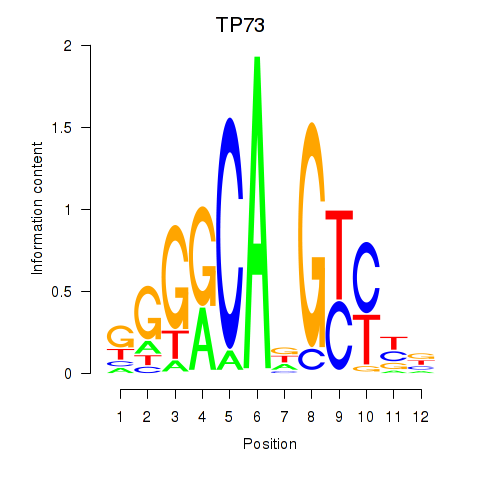
Activity profile of TP73 motif
Sorted Z-values of TP73 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TP73
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 1.2 | 4.9 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 1.2 | 10.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.5 | 1.5 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.4 | 2.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 1.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 8.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 0.8 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 0.9 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 2.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 1.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.9 | GO:0046469 | plasma lipoprotein particle oxidation(GO:0034441) platelet activating factor metabolic process(GO:0046469) |
| 0.1 | 1.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.4 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.1 | 2.1 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 2.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.9 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 0.9 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.6 | GO:0034378 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) chylomicron assembly(GO:0034378) positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.6 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 1.8 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 1.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0014735 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 1.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 2.1 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.0 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.0 | 0.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.0 | 4.9 | GO:0044393 | microspike(GO:0044393) |
| 0.7 | 2.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.2 | 0.6 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 2.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 1.2 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.3 | 0.9 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 2.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 1.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 2.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 10.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 0.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 1.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.6 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 4.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 8.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 2.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 2.1 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |