Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
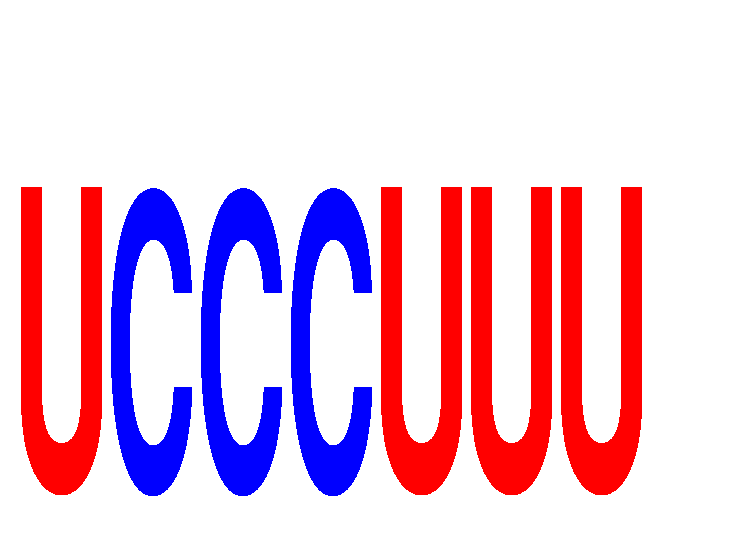
Results for UCCCUUU
Z-value: 0.28
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-204-5p
|
MIMAT0000265 |
|
hsa-miR-211-5p
|
MIMAT0000268 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.0 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.0 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.0 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.0 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |