Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
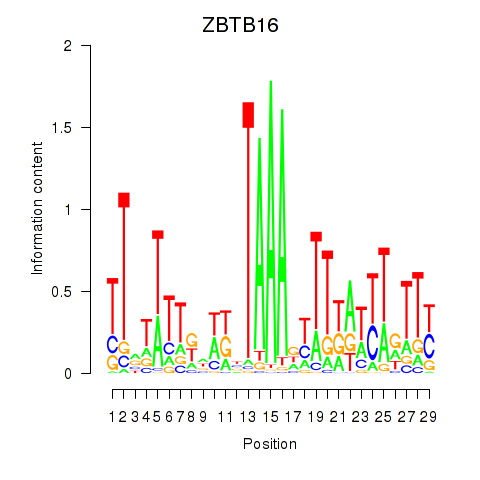
Results for ZBTB16
Z-value: 1.89
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB16 | hg19_v2_chr11_+_113930291_113930339 | -0.75 | 1.6e-04 | Click! |
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.7 | 4.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.6 | 1.9 | GO:0018013 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.5 | 2.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.5 | 1.9 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.5 | 3.3 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.5 | 2.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.5 | 4.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 2.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 1.5 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.3 | 1.4 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.3 | 1.7 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.3 | 1.0 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.3 | 2.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 1.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 1.8 | GO:0035701 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.3 | 2.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.3 | 5.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 1.6 | GO:0015853 | adenine transport(GO:0015853) |
| 0.2 | 1.7 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 1.6 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.2 | 2.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.6 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 2.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 1.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 2.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.5 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.1 | 0.5 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 1.7 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.4 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 1.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 5.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 1.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.7 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.5 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 2.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.3 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.7 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 2.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 2.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 2.0 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 2.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.5 | GO:0009304 | transcription initiation from RNA polymerase III promoter(GO:0006384) tRNA transcription(GO:0009304) |
| 0.0 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 2.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 2.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 3.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.8 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 3.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.8 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 3.7 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.5 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.9 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 1.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.6 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 1.5 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.4 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 1.5 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 1.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.5 | 1.5 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.4 | 1.4 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.4 | 2.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 1.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 2.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 1.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 2.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 1.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 2.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 2.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 6.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.0 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.8 | 2.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.8 | 2.3 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.7 | 2.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 1.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.4 | 1.8 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.4 | 2.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.4 | 1.6 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.4 | 1.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 1.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 5.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 2.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 3.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 3.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.5 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 4.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 2.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 2.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 3.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 3.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 4.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 4.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 3.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 4.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.0 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 4.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 5.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.0 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 5.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |