Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
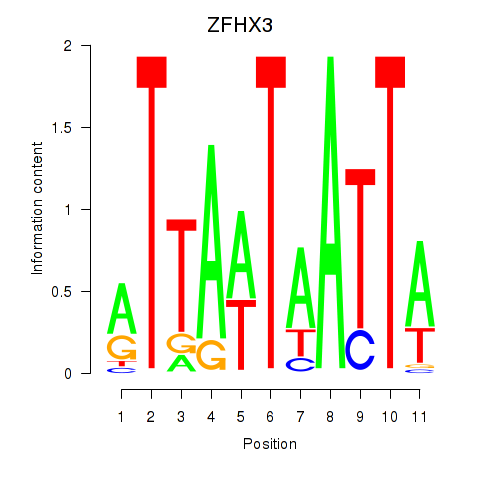
Results for ZFHX3
Z-value: 0.87
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.10 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFHX3 | hg19_v2_chr16_-_73093597_73093597 | 0.25 | 2.9e-01 | Click! |
Activity profile of ZFHX3 motif
Sorted Z-values of ZFHX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFHX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 1.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 1.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 0.8 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.2 | 3.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.0 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.2 | 1.9 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.5 | GO:2000007 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 1.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.4 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 1.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0061364 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 1.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 3.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 1.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 1.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 3.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 0.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.8 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.4 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.1 | 1.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 1.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 1.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.4 | GO:0070492 | phosphatidylinositol-5-phosphate binding(GO:0010314) oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |