Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
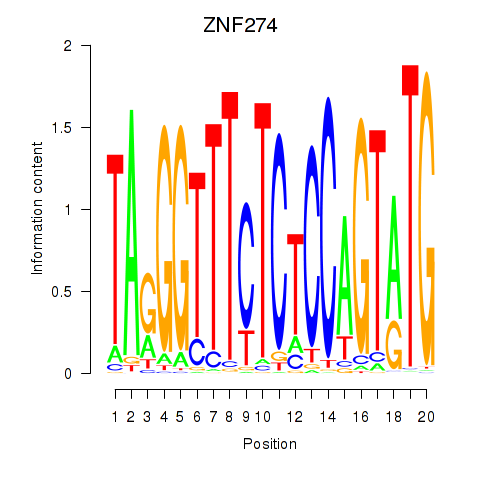
Results for ZNF274
Z-value: 1.00
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF274 | hg19_v2_chr19_+_58694396_58694485 | -0.25 | 2.8e-01 | Click! |
Activity profile of ZNF274 motif
Sorted Z-values of ZNF274 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF274
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 2.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.0 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 2.5 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.3 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 2.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.7 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.5 | 3.6 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 2.7 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 1.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 3.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |