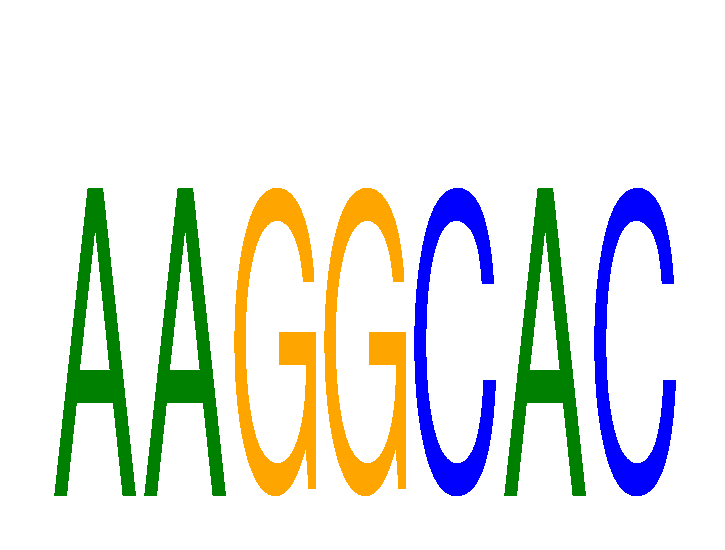|
chr1_-_242524687
Show fit
|
0.67 |
ENST00000442594.6
ENST00000536534.7
|
PLD5
|
phospholipase D family member 5
|
|
chr14_-_105168753
Show fit
|
0.63 |
ENST00000331782.8
ENST00000347004.2
|
JAG2
|
jagged canonical Notch ligand 2
|
|
chr1_+_92029971
Show fit
|
0.54 |
ENST00000370383.5
|
EPHX4
|
epoxide hydrolase 4
|
|
chr7_+_121873152
Show fit
|
0.53 |
ENST00000650826.1
ENST00000650728.1
ENST00000393386.7
ENST00000651390.1
ENST00000651842.1
ENST00000650681.1
|
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1
|
|
chr1_+_151511376
Show fit
|
0.51 |
ENST00000427934.2
ENST00000271636.12
|
CGN
|
cingulin
|
|
chr9_+_113150991
Show fit
|
0.47 |
ENST00000259392.8
|
SLC31A2
|
solute carrier family 31 member 2
|
|
chr1_-_161021096
Show fit
|
0.47 |
ENST00000537746.1
ENST00000368026.11
|
F11R
|
F11 receptor
|
|
chr18_+_22169580
Show fit
|
0.46 |
ENST00000269216.10
|
GATA6
|
GATA binding protein 6
|
|
chr3_-_69386079
Show fit
|
0.44 |
ENST00000398540.8
|
FRMD4B
|
FERM domain containing 4B
|
|
chr10_+_93893931
Show fit
|
0.44 |
ENST00000371408.7
ENST00000427197.2
|
SLC35G1
|
solute carrier family 35 member G1
|
|
chrX_+_106693838
Show fit
|
0.43 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128
|
|
chr20_-_14337602
Show fit
|
0.43 |
ENST00000378053.3
ENST00000341420.5
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr8_-_15238423
Show fit
|
0.42 |
ENST00000382080.6
|
SGCZ
|
sarcoglycan zeta
|
|
chr1_+_233904656
Show fit
|
0.41 |
ENST00000366618.8
|
SLC35F3
|
solute carrier family 35 member F3
|
|
chr2_+_17541157
Show fit
|
0.40 |
ENST00000406397.1
|
VSNL1
|
visinin like 1
|
|
chr5_-_16508990
Show fit
|
0.39 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1
|
|
chr14_-_99272184
Show fit
|
0.39 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B
|
|
chr10_-_103855406
Show fit
|
0.37 |
ENST00000355946.6
ENST00000369774.8
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr11_-_115504389
Show fit
|
0.37 |
ENST00000545380.5
ENST00000452722.7
ENST00000331581.11
ENST00000537058.5
ENST00000536727.5
ENST00000542447.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr6_-_136550407
Show fit
|
0.36 |
ENST00000354570.8
|
MAP7
|
microtubule associated protein 7
|
|
chr19_+_53867874
Show fit
|
0.36 |
ENST00000448420.5
ENST00000439000.5
ENST00000391771.1
ENST00000391770.9
|
MYADM
|
myeloid associated differentiation marker
|
|
chr4_-_80073170
Show fit
|
0.36 |
ENST00000403729.7
|
ANTXR2
|
ANTXR cell adhesion molecule 2
|
|
chr6_+_1312090
Show fit
|
0.36 |
ENST00000296839.5
|
FOXQ1
|
forkhead box Q1
|
|
chr20_-_18057841
Show fit
|
0.35 |
ENST00000278780.7
|
OVOL2
|
ovo like zinc finger 2
|
|
chr12_-_94650506
Show fit
|
0.35 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr14_-_69574845
Show fit
|
0.35 |
ENST00000599174.3
|
CCDC177
|
coiled-coil domain containing 177
|
|
chr15_-_78944985
Show fit
|
0.34 |
ENST00000615999.5
ENST00000677789.1
ENST00000676880.1
ENST00000677936.1
ENST00000220166.10
ENST00000677810.1
ENST00000678644.1
ENST00000677534.1
ENST00000677316.1
|
CTSH
|
cathepsin H
|
|
chr2_-_121285194
Show fit
|
0.34 |
ENST00000263707.6
|
TFCP2L1
|
transcription factor CP2 like 1
|
|
chr1_-_201399302
Show fit
|
0.33 |
ENST00000633953.1
ENST00000391967.7
|
LAD1
|
ladinin 1
|
|
chr2_+_26346086
Show fit
|
0.33 |
ENST00000613142.4
ENST00000260585.12
ENST00000447170.1
|
SELENOI
|
selenoprotein I
|
|
chr11_+_10450289
Show fit
|
0.32 |
ENST00000444303.6
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr18_+_11689210
Show fit
|
0.32 |
ENST00000334049.11
|
GNAL
|
G protein subunit alpha L
|
|
chr9_+_470291
Show fit
|
0.31 |
ENST00000382303.5
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr2_+_112055201
Show fit
|
0.31 |
ENST00000283206.9
|
TMEM87B
|
transmembrane protein 87B
|
|
chr14_+_67533282
Show fit
|
0.30 |
ENST00000329153.10
|
PLEKHH1
|
pleckstrin homology, MyTH4 and FERM domain containing H1
|
|
chr2_-_207769889
Show fit
|
0.29 |
ENST00000295417.4
|
FZD5
|
frizzled class receptor 5
|
|
chr18_-_50195138
Show fit
|
0.29 |
ENST00000285039.12
|
MYO5B
|
myosin VB
|
|
chr7_-_108456378
Show fit
|
0.29 |
ENST00000613830.4
ENST00000413765.6
ENST00000379028.8
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr6_+_7726089
Show fit
|
0.29 |
ENST00000283147.7
|
BMP6
|
bone morphogenetic protein 6
|
|
chr10_-_35642286
Show fit
|
0.29 |
ENST00000374694.3
|
FZD8
|
frizzled class receptor 8
|
|
chr8_-_143939543
Show fit
|
0.29 |
ENST00000345136.8
|
PLEC
|
plectin
|
|
chr22_+_28883564
Show fit
|
0.28 |
ENST00000544604.7
|
ZNRF3
|
zinc and ring finger 3
|
|
chr12_-_104958268
Show fit
|
0.28 |
ENST00000432951.1
ENST00000258538.8
ENST00000415674.1
ENST00000424946.1
ENST00000433540.5
|
SLC41A2
|
solute carrier family 41 member 2
|
|
chr16_-_17470953
Show fit
|
0.27 |
ENST00000261381.7
|
XYLT1
|
xylosyltransferase 1
|
|
chr2_-_96740034
Show fit
|
0.27 |
ENST00000264963.9
ENST00000377079.8
|
LMAN2L
|
lectin, mannose binding 2 like
|
|
chr4_+_94757921
Show fit
|
0.27 |
ENST00000515059.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr14_+_64704380
Show fit
|
0.27 |
ENST00000247226.13
ENST00000394691.7
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3
|
|
chr15_+_33310946
Show fit
|
0.26 |
ENST00000415757.7
ENST00000634891.2
ENST00000389232.9
ENST00000622037.1
|
RYR3
|
ryanodine receptor 3
|
|
chr1_+_31413187
Show fit
|
0.26 |
ENST00000373709.8
|
SERINC2
|
serine incorporator 2
|
|
chr9_+_22446808
Show fit
|
0.25 |
ENST00000325870.3
|
DMRTA1
|
DMRT like family A1
|
|
chr19_+_38433676
Show fit
|
0.25 |
ENST00000359596.8
ENST00000355481.8
|
RYR1
|
ryanodine receptor 1
|
|
chr19_-_10587219
Show fit
|
0.25 |
ENST00000591240.5
ENST00000589684.5
ENST00000591676.1
ENST00000250244.11
ENST00000590923.5
|
AP1M2
|
adaptor related protein complex 1 subunit mu 2
|
|
chr12_+_40692413
Show fit
|
0.25 |
ENST00000551295.7
ENST00000547702.5
ENST00000551424.5
|
CNTN1
|
contactin 1
|
|
chr12_+_65169546
Show fit
|
0.25 |
ENST00000308330.3
|
LEMD3
|
LEM domain containing 3
|
|
chr3_-_185498964
Show fit
|
0.24 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A
|
|
chr12_+_4273751
Show fit
|
0.24 |
ENST00000675880.1
ENST00000261254.8
|
CCND2
|
cyclin D2
|
|
chr2_-_9003657
Show fit
|
0.24 |
ENST00000462696.1
ENST00000305997.8
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr1_+_26529745
Show fit
|
0.24 |
ENST00000374168.7
ENST00000374166.8
|
RPS6KA1
|
ribosomal protein S6 kinase A1
|
|
chr8_+_60678705
Show fit
|
0.24 |
ENST00000423902.7
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr8_+_28701487
Show fit
|
0.24 |
ENST00000220562.9
|
EXTL3
|
exostosin like glycosyltransferase 3
|
|
chr14_+_69611586
Show fit
|
0.24 |
ENST00000342745.5
|
SUSD6
|
sushi domain containing 6
|
|
chr18_+_32018817
Show fit
|
0.24 |
ENST00000217740.4
ENST00000583184.1
|
RNF125
ENSG00000263917.1
|
ring finger protein 125
novel transcript
|
|
chrX_+_68829009
Show fit
|
0.23 |
ENST00000204961.5
|
EFNB1
|
ephrin B1
|
|
chr1_-_115768702
Show fit
|
0.23 |
ENST00000261448.6
|
CASQ2
|
calsequestrin 2
|
|
chr6_+_12012304
Show fit
|
0.23 |
ENST00000379388.7
ENST00000627968.2
ENST00000541134.5
|
HIVEP1
|
HIVEP zinc finger 1
|
|
chr11_+_118359572
Show fit
|
0.23 |
ENST00000252108.8
ENST00000431736.6
|
UBE4A
|
ubiquitination factor E4A
|
|
chr9_-_133121228
Show fit
|
0.23 |
ENST00000372050.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr15_-_59372863
Show fit
|
0.22 |
ENST00000288235.9
|
MYO1E
|
myosin IE
|
|
chr9_+_98807619
Show fit
|
0.22 |
ENST00000375011.4
|
GALNT12
|
polypeptide N-acetylgalactosaminyltransferase 12
|
|
chr10_-_15371225
Show fit
|
0.22 |
ENST00000378116.9
|
FAM171A1
|
family with sequence similarity 171 member A1
|
|
chr6_+_33620329
Show fit
|
0.22 |
ENST00000374316.9
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor type 3
|
|
chr9_-_121370235
Show fit
|
0.22 |
ENST00000286713.7
ENST00000347359.3
|
STOM
|
stomatin
|
|
chr6_+_37170133
Show fit
|
0.22 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase
|
|
chr13_+_31199959
Show fit
|
0.22 |
ENST00000343307.5
|
B3GLCT
|
beta 3-glucosyltransferase
|
|
chr18_-_21704763
Show fit
|
0.22 |
ENST00000580981.5
ENST00000289119.7
|
ABHD3
|
abhydrolase domain containing 3, phospholipase
|
|
chr6_+_44219595
Show fit
|
0.21 |
ENST00000393844.7
ENST00000652680.1
ENST00000643028.2
ENST00000371713.6
|
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group)
|
|
chr2_+_69915100
Show fit
|
0.21 |
ENST00000264444.7
|
MXD1
|
MAX dimerization protein 1
|
|
chr9_-_10612966
Show fit
|
0.21 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D
|
|
chr17_-_49764123
Show fit
|
0.21 |
ENST00000240364.7
ENST00000506156.1
|
FAM117A
|
family with sequence similarity 117 member A
|
|
chr5_-_157575767
Show fit
|
0.21 |
ENST00000257527.9
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr5_+_140841183
Show fit
|
0.21 |
ENST00000378123.4
ENST00000531613.2
|
PCDHA8
|
protocadherin alpha 8
|
|
chr20_-_57710001
Show fit
|
0.20 |
ENST00000341744.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr3_+_5187697
Show fit
|
0.20 |
ENST00000256497.9
|
EDEM1
|
ER degradation enhancing alpha-mannosidase like protein 1
|
|
chr10_+_69451456
Show fit
|
0.20 |
ENST00000373290.7
|
TSPAN15
|
tetraspanin 15
|
|
chr1_-_226737277
Show fit
|
0.20 |
ENST00000272117.7
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr8_+_73294594
Show fit
|
0.20 |
ENST00000240285.10
|
RDH10
|
retinol dehydrogenase 10
|
|
chr5_+_140875299
Show fit
|
0.20 |
ENST00000613593.1
ENST00000398631.3
|
PCDHA12
|
protocadherin alpha 12
|
|
chr11_+_121452291
Show fit
|
0.20 |
ENST00000260197.12
|
SORL1
|
sortilin related receptor 1
|
|
chr2_+_118088432
Show fit
|
0.20 |
ENST00000245787.9
|
INSIG2
|
insulin induced gene 2
|
|
chr2_+_205682491
Show fit
|
0.20 |
ENST00000360409.7
ENST00000450507.5
ENST00000357785.10
ENST00000417189.5
|
NRP2
|
neuropilin 2
|
|
chr5_+_140848360
Show fit
|
0.20 |
ENST00000532602.2
|
PCDHA9
|
protocadherin alpha 9
|
|
chr5_+_140868945
Show fit
|
0.20 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11
|
|
chr12_+_43836043
Show fit
|
0.20 |
ENST00000266534.8
ENST00000551577.5
|
TMEM117
|
transmembrane protein 117
|
|
chr12_+_4909895
Show fit
|
0.19 |
ENST00000638821.1
ENST00000382545.5
|
ENSG00000256654.4
KCNA1
|
novel transcript, sense overlapping KCNA1
potassium voltage-gated channel subfamily A member 1
|
|
chr16_-_2196575
Show fit
|
0.19 |
ENST00000343516.8
|
CASKIN1
|
CASK interacting protein 1
|
|
chr11_+_76783349
Show fit
|
0.19 |
ENST00000333090.5
|
TSKU
|
tsukushi, small leucine rich proteoglycan
|
|
chr12_-_122896066
Show fit
|
0.19 |
ENST00000267202.7
ENST00000535765.5
|
VPS37B
|
VPS37B subunit of ESCRT-I
|
|
chr20_-_51802509
Show fit
|
0.19 |
ENST00000371539.7
ENST00000217086.9
|
SALL4
|
spalt like transcription factor 4
|
|
chr5_+_128083757
Show fit
|
0.19 |
ENST00000262461.7
ENST00000628403.2
ENST00000343225.4
|
SLC12A2
|
solute carrier family 12 member 2
|
|
chr7_-_101217569
Show fit
|
0.19 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3
|
|
chr11_+_65787056
Show fit
|
0.19 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1
|
|
chr8_-_74321532
Show fit
|
0.19 |
ENST00000342232.5
|
JPH1
|
junctophilin 1
|
|
chr14_-_23183641
Show fit
|
0.19 |
ENST00000469263.5
ENST00000525062.1
ENST00000316902.12
ENST00000524758.1
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr14_-_77616630
Show fit
|
0.19 |
ENST00000216484.7
|
SPTLC2
|
serine palmitoyltransferase long chain base subunit 2
|
|
chr16_-_68448491
Show fit
|
0.19 |
ENST00000561749.1
ENST00000219334.10
|
SMPD3
|
sphingomyelin phosphodiesterase 3
|
|
chr21_-_44873671
Show fit
|
0.19 |
ENST00000330938.8
ENST00000397887.7
|
PTTG1IP
|
PTTG1 interacting protein
|
|
chr1_+_1020068
Show fit
|
0.19 |
ENST00000379370.7
ENST00000620552.4
|
AGRN
|
agrin
|
|
chr1_-_18956669
Show fit
|
0.19 |
ENST00000455833.7
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr9_+_2621766
Show fit
|
0.18 |
ENST00000382100.8
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr11_+_60914139
Show fit
|
0.18 |
ENST00000227525.8
|
TMEM109
|
transmembrane protein 109
|
|
chr19_-_55443263
Show fit
|
0.18 |
ENST00000416792.2
ENST00000376325.10
|
SHISA7
|
shisa family member 7
|
|
chr10_+_86756580
Show fit
|
0.18 |
ENST00000372037.8
|
BMPR1A
|
bone morphogenetic protein receptor type 1A
|
|
chr1_+_65147514
Show fit
|
0.18 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4
|
|
chr10_-_50623897
Show fit
|
0.18 |
ENST00000361781.7
ENST00000429490.5
ENST00000619438.4
|
SGMS1
|
sphingomyelin synthase 1
|
|
chr11_+_120210991
Show fit
|
0.18 |
ENST00000328965.9
|
OAF
|
out at first homolog
|
|
chr1_+_40258202
Show fit
|
0.18 |
ENST00000372759.4
|
ZMPSTE24
|
zinc metallopeptidase STE24
|
|
chr16_+_55509006
Show fit
|
0.18 |
ENST00000262134.10
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2
|
|
chr3_-_4467241
Show fit
|
0.18 |
ENST00000383843.9
ENST00000272902.10
ENST00000458465.6
ENST00000405420.2
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr3_+_36380477
Show fit
|
0.18 |
ENST00000457375.6
ENST00000273183.8
ENST00000434649.1
|
STAC
|
SH3 and cysteine rich domain
|
|
chr14_-_95319863
Show fit
|
0.18 |
ENST00000298912.9
|
CLMN
|
calmin
|
|
chr8_+_96261891
Show fit
|
0.17 |
ENST00000517309.6
|
PTDSS1
|
phosphatidylserine synthase 1
|
|
chr1_+_168225950
Show fit
|
0.17 |
ENST00000367829.5
ENST00000271375.7
ENST00000630869.1
|
SFT2D2
|
SFT2 domain containing 2
|
|
chr12_-_128823942
Show fit
|
0.17 |
ENST00000266771.10
|
SLC15A4
|
solute carrier family 15 member 4
|
|
chr11_+_7576408
Show fit
|
0.17 |
ENST00000533792.5
|
PPFIBP2
|
PPFIA binding protein 2
|
|
chr19_-_45405034
Show fit
|
0.17 |
ENST00000592134.1
ENST00000360957.10
|
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like
|
|
chr13_+_42048645
Show fit
|
0.17 |
ENST00000337343.9
ENST00000261491.9
ENST00000611224.1
|
DGKH
|
diacylglycerol kinase eta
|
|
chr1_+_25430854
Show fit
|
0.17 |
ENST00000399766.7
|
MACO1
|
macoilin 1
|
|
chr2_+_119223815
Show fit
|
0.17 |
ENST00000393106.6
ENST00000393110.7
ENST00000409811.5
ENST00000393107.2
|
STEAP3
|
STEAP3 metalloreductase
|
|
chr8_-_56993803
Show fit
|
0.17 |
ENST00000262644.9
|
BPNT2
|
3'(2'), 5'-bisphosphate nucleotidase 2
|
|
chr9_-_34637719
Show fit
|
0.17 |
ENST00000378892.5
ENST00000680277.1
ENST00000277010.9
ENST00000679597.1
ENST00000680244.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1
|
|
chr20_-_45348414
Show fit
|
0.17 |
ENST00000372733.3
|
SDC4
|
syndecan 4
|
|
chr17_+_3636749
Show fit
|
0.17 |
ENST00000046640.9
ENST00000574218.1
ENST00000399306.7
|
CTNS
|
cystinosin, lysosomal cystine transporter
|
|
chr11_-_88175432
Show fit
|
0.16 |
ENST00000531138.1
ENST00000526372.1
ENST00000243662.11
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr9_-_34376878
Show fit
|
0.16 |
ENST00000297625.8
|
MYORG
|
myogenesis regulating glycosidase (putative)
|
|
chr10_+_70815889
Show fit
|
0.16 |
ENST00000373202.8
|
SGPL1
|
sphingosine-1-phosphate lyase 1
|
|
chr21_+_17513119
Show fit
|
0.16 |
ENST00000356275.10
ENST00000400165.5
ENST00000400169.1
|
CXADR
|
CXADR Ig-like cell adhesion molecule
|
|
chr22_-_50307598
Show fit
|
0.16 |
ENST00000425954.1
ENST00000449103.5
ENST00000359337.9
|
PLXNB2
|
plexin B2
|
|
chr13_+_79481124
Show fit
|
0.16 |
ENST00000612570.4
ENST00000218652.11
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr8_+_97869040
Show fit
|
0.16 |
ENST00000254898.7
ENST00000524308.5
ENST00000522025.6
|
MATN2
|
matrilin 2
|
|
chr17_-_17591658
Show fit
|
0.16 |
ENST00000435340.6
ENST00000255389.10
ENST00000395781.6
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr7_-_113087720
Show fit
|
0.16 |
ENST00000297146.7
|
GPR85
|
G protein-coupled receptor 85
|
|
chr6_+_72622047
Show fit
|
0.16 |
ENST00000370398.6
|
KCNQ5
|
potassium voltage-gated channel subfamily Q member 5
|
|
chr6_+_7107941
Show fit
|
0.16 |
ENST00000379938.7
ENST00000467782.5
ENST00000334984.10
ENST00000349384.10
|
RREB1
|
ras responsive element binding protein 1
|
|
chr19_+_38647614
Show fit
|
0.16 |
ENST00000252699.7
ENST00000424234.7
ENST00000440400.2
|
ACTN4
|
actinin alpha 4
|
|
chr19_-_55370455
Show fit
|
0.16 |
ENST00000264563.7
ENST00000585513.1
ENST00000590625.5
|
IL11
|
interleukin 11
|
|
chr15_-_48963912
Show fit
|
0.16 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4
|
|
chr4_+_151409169
Show fit
|
0.15 |
ENST00000435205.6
|
FAM160A1
|
family with sequence similarity 160 member A1
|
|
chr12_-_57078739
Show fit
|
0.15 |
ENST00000379391.7
|
NEMP1
|
nuclear envelope integral membrane protein 1
|
|
chr11_+_20599602
Show fit
|
0.15 |
ENST00000525748.6
|
SLC6A5
|
solute carrier family 6 member 5
|
|
chr8_+_97644164
Show fit
|
0.15 |
ENST00000336273.8
|
MTDH
|
metadherin
|
|
chr15_-_50765656
Show fit
|
0.15 |
ENST00000261854.10
|
SPPL2A
|
signal peptide peptidase like 2A
|
|
chr12_-_7018465
Show fit
|
0.15 |
ENST00000261407.9
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3
|
|
chr1_-_112956063
Show fit
|
0.15 |
ENST00000538576.5
ENST00000369626.8
ENST00000458229.6
|
SLC16A1
|
solute carrier family 16 member 1
|
|
chr2_+_11155498
Show fit
|
0.15 |
ENST00000402361.5
ENST00000428481.1
|
SLC66A3
|
solute carrier family 66 member 3
|
|
chr12_+_113221429
Show fit
|
0.15 |
ENST00000551096.5
ENST00000551099.5
ENST00000552897.5
ENST00000550785.5
ENST00000549279.1
ENST00000335509.11
|
TPCN1
|
two pore segment channel 1
|
|
chr5_+_109689915
Show fit
|
0.15 |
ENST00000261483.5
|
MAN2A1
|
mannosidase alpha class 2A member 1
|
|
chr1_+_115641945
Show fit
|
0.15 |
ENST00000355485.7
ENST00000369510.8
|
VANGL1
|
VANGL planar cell polarity protein 1
|
|
chr7_-_158829519
Show fit
|
0.15 |
ENST00000251527.10
ENST00000652148.1
|
ESYT2
|
extended synaptotagmin 2
|
|
chr1_-_169485931
Show fit
|
0.15 |
ENST00000367804.4
ENST00000646596.1
ENST00000236137.10
|
SLC19A2
|
solute carrier family 19 member 2
|
|
chr14_+_70641896
Show fit
|
0.15 |
ENST00000256367.3
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr18_-_55588184
Show fit
|
0.15 |
ENST00000354452.8
ENST00000565908.6
ENST00000635822.2
|
TCF4
|
transcription factor 4
|
|
chr7_-_105522204
Show fit
|
0.15 |
ENST00000356362.6
|
PUS7
|
pseudouridine synthase 7
|
|
chr7_-_112790372
Show fit
|
0.15 |
ENST00000449743.1
ENST00000441474.1
ENST00000312814.11
ENST00000454074.5
ENST00000447395.5
|
TMEM168
|
transmembrane protein 168
|
|
chr6_-_33789090
Show fit
|
0.14 |
ENST00000614475.4
ENST00000293760.10
|
LEMD2
|
LEM domain nuclear envelope protein 2
|
|
chr8_-_53251585
Show fit
|
0.14 |
ENST00000265572.8
ENST00000673285.2
|
OPRK1
|
opioid receptor kappa 1
|
|
chr9_+_113876282
Show fit
|
0.14 |
ENST00000374126.9
ENST00000615615.4
ENST00000288466.11
|
ZNF618
|
zinc finger protein 618
|
|
chr6_-_43575966
Show fit
|
0.14 |
ENST00000265351.12
|
XPO5
|
exportin 5
|
|
chr3_-_27456743
Show fit
|
0.14 |
ENST00000295736.9
ENST00000428386.5
ENST00000428179.1
|
SLC4A7
|
solute carrier family 4 member 7
|
|
chrX_-_33128360
Show fit
|
0.14 |
ENST00000378677.6
|
DMD
|
dystrophin
|
|
chr12_-_92929236
Show fit
|
0.14 |
ENST00000322349.13
|
EEA1
|
early endosome antigen 1
|
|
chr22_-_36387949
Show fit
|
0.14 |
ENST00000216181.11
|
MYH9
|
myosin heavy chain 9
|
|
chr7_+_77537258
Show fit
|
0.14 |
ENST00000248594.11
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12
|
|
chr5_+_140827950
Show fit
|
0.14 |
ENST00000378126.4
ENST00000529310.6
ENST00000527624.1
|
PCDHA6
|
protocadherin alpha 6
|
|
chr12_+_123712333
Show fit
|
0.14 |
ENST00000330342.8
ENST00000613625.5
|
ATP6V0A2
|
ATPase H+ transporting V0 subunit a2
|
|
chr12_-_89352487
Show fit
|
0.14 |
ENST00000548755.1
ENST00000279488.8
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr4_+_107824555
Show fit
|
0.14 |
ENST00000394684.8
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr6_+_53794948
Show fit
|
0.14 |
ENST00000370888.6
|
LRRC1
|
leucine rich repeat containing 1
|
|
chr2_-_113756628
Show fit
|
0.14 |
ENST00000245680.7
|
SLC35F5
|
solute carrier family 35 member F5
|
|
chr9_-_109119915
Show fit
|
0.14 |
ENST00000374586.8
|
TMEM245
|
transmembrane protein 245
|
|
chr11_-_72080389
Show fit
|
0.14 |
ENST00000351960.10
ENST00000541719.5
ENST00000535111.5
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr11_+_69048889
Show fit
|
0.14 |
ENST00000294309.8
ENST00000637504.1
ENST00000542467.1
ENST00000637342.1
|
TPCN2
|
two pore segment channel 2
|
|
chr1_+_220528112
Show fit
|
0.14 |
ENST00000366917.6
ENST00000402574.5
ENST00000611084.4
ENST00000366918.8
|
MARK1
|
microtubule affinity regulating kinase 1
|
|
chr11_-_59615673
Show fit
|
0.14 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein
|
|
chr1_+_100896060
Show fit
|
0.14 |
ENST00000370112.8
ENST00000357650.9
|
SLC30A7
|
solute carrier family 30 member 7
|
|
chr12_+_52010485
Show fit
|
0.14 |
ENST00000552049.5
ENST00000546756.1
|
TAMALIN
|
trafficking regulator and scaffold protein tamalin
|
|
chr6_-_75206044
Show fit
|
0.14 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain
|
|
chr5_+_56815534
Show fit
|
0.14 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr10_-_104338431
Show fit
|
0.13 |
ENST00000647721.1
ENST00000337478.3
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein
|
|
chr6_+_149566356
Show fit
|
0.13 |
ENST00000367419.10
|
GINM1
|
glycoprotein integral membrane 1
|
|
chr5_-_16936231
Show fit
|
0.13 |
ENST00000507288.1
ENST00000274203.13
ENST00000513610.6
|
MYO10
|
myosin X
|
|
chr14_-_63728027
Show fit
|
0.13 |
ENST00000247225.7
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1
|
|
chr13_-_29595670
Show fit
|
0.13 |
ENST00000380752.10
|
SLC7A1
|
solute carrier family 7 member 1
|
|
chr1_+_99969979
Show fit
|
0.13 |
ENST00000427993.7
ENST00000533028.8
ENST00000639221.1
ENST00000638336.1
ENST00000639807.1
ENST00000640715.1
ENST00000465289.6
ENST00000639994.1
ENST00000638988.1
ENST00000640732.1
ENST00000640600.1
ENST00000638338.1
ENST00000638792.1
ENST00000639037.1
|
SLC35A3
ENSG00000283761.1
|
solute carrier family 35 member A3
novel protein
|
|
chr11_+_75768769
Show fit
|
0.13 |
ENST00000228027.12
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr8_-_65842051
Show fit
|
0.13 |
ENST00000401827.8
|
PDE7A
|
phosphodiesterase 7A
|
|
chr5_+_149730260
Show fit
|
0.13 |
ENST00000360453.8
ENST00000394320.7
ENST00000309241.10
|
PPARGC1B
|
PPARG coactivator 1 beta
|
|
chr9_-_99221897
Show fit
|
0.13 |
ENST00000476832.2
|
ALG2
|
ALG2 alpha-1,3/1,6-mannosyltransferase
|
|
chr22_+_21642287
Show fit
|
0.13 |
ENST00000248958.5
|
SDF2L1
|
stromal cell derived factor 2 like 1
|
|
chr5_+_140966466
Show fit
|
0.13 |
ENST00000615316.1
ENST00000289269.7
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr4_+_165873231
Show fit
|
0.13 |
ENST00000061240.7
|
TLL1
|
tolloid like 1
|