Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
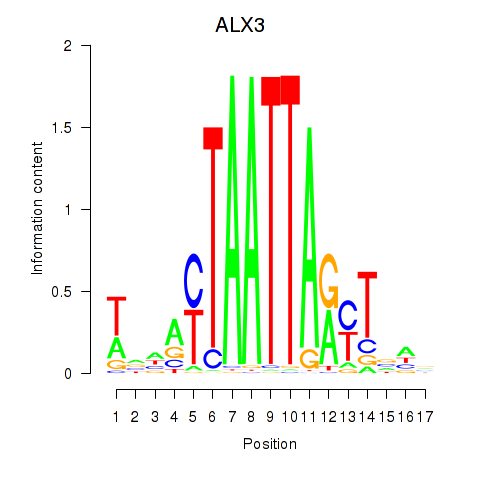
Results for ALX3
Z-value: 1.02
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.9 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg38_v1_chr1_-_110070654_110070677 | 0.74 | 3.6e-02 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_104085847 | 2.10 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr18_+_31447732 | 1.69 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr7_+_70596078 | 1.51 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr3_-_151316795 | 1.44 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr1_+_27934980 | 1.40 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr1_+_27935110 | 1.30 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr17_-_7263959 | 1.20 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr1_+_160400543 | 1.18 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr2_+_68734773 | 1.09 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr5_+_67004618 | 0.90 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_24466487 | 0.86 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr9_-_114348966 | 0.85 |
ENST00000374079.8
|
AKNA
|
AT-hook transcription factor |
| chr9_-_5304713 | 0.80 |
ENST00000381627.4
|
RLN2
|
relaxin 2 |
| chr8_-_85341705 | 0.79 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr2_+_68734861 | 0.76 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr3_-_191282383 | 0.75 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr6_+_130421086 | 0.72 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A |
| chr6_+_132538290 | 0.71 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr13_-_85799400 | 0.69 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr16_-_28623560 | 0.67 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr1_+_27935022 | 0.66 |
ENST00000411604.5
ENST00000373888.8 |
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr7_-_111392915 | 0.65 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chrX_+_136536099 | 0.63 |
ENST00000440515.5
ENST00000456412.1 |
VGLL1
|
vestigial like family member 1 |
| chr4_-_73620391 | 0.63 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr1_+_81306096 | 0.58 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr14_-_67412112 | 0.57 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr4_-_25863537 | 0.56 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr3_-_112829367 | 0.56 |
ENST00000448932.4
ENST00000617549.3 |
CD200R1L
|
CD200 receptor 1 like |
| chr8_+_106726115 | 0.52 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr8_+_104223320 | 0.52 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_+_130018565 | 0.51 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr3_-_142029108 | 0.51 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr1_-_99766620 | 0.50 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr9_-_5339874 | 0.48 |
ENST00000223862.2
|
RLN1
|
relaxin 1 |
| chr20_-_52105644 | 0.47 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr16_+_11965193 | 0.46 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr11_-_5301946 | 0.44 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr1_-_247760556 | 0.43 |
ENST00000641256.1
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr8_+_104223344 | 0.42 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_+_108045050 | 0.41 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_+_6876625 | 0.41 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor family 10 subfamily A member 4 |
| chr5_+_140806929 | 0.40 |
ENST00000378125.4
ENST00000618834.1 ENST00000530339.2 ENST00000512229.6 ENST00000672575.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr4_-_73620629 | 0.40 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr8_-_85341659 | 0.40 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr8_+_49911801 | 0.40 |
ENST00000643809.1
|
SNTG1
|
syntrophin gamma 1 |
| chr5_-_140346596 | 0.40 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr3_-_161105399 | 0.39 |
ENST00000652593.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr6_-_53510445 | 0.37 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chr3_-_161105224 | 0.37 |
ENST00000651254.1
ENST00000651178.1 ENST00000476999.6 ENST00000652596.1 ENST00000651305.1 ENST00000652111.1 ENST00000651292.1 ENST00000651282.1 ENST00000651380.1 ENST00000494173.7 ENST00000484127.5 ENST00000650733.1 ENST00000494818.6 ENST00000492353.5 ENST00000652143.1 ENST00000473142.5 ENST00000651147.1 ENST00000468268.5 ENST00000460353.2 ENST00000651953.1 ENST00000651972.1 ENST00000652730.1 ENST00000651460.1 ENST00000652059.1 ENST00000651509.1 ENST00000651801.1 ENST00000651686.1 ENST00000320474.10 ENST00000392781.7 ENST00000392779.6 ENST00000651791.1 ENST00000651117.1 ENST00000652032.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr8_-_90082871 | 0.36 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr20_+_33217325 | 0.36 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chrX_+_108044967 | 0.36 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr9_+_470291 | 0.35 |
ENST00000382303.5
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr17_+_37491464 | 0.35 |
ENST00000613659.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr3_-_161105070 | 0.34 |
ENST00000651430.1
ENST00000650695.1 ENST00000651689.1 ENST00000651916.1 ENST00000488170.5 ENST00000652377.1 ENST00000652669.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr8_+_106726012 | 0.34 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr18_-_55423757 | 0.34 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chr2_-_101308681 | 0.34 |
ENST00000295317.4
|
RNF149
|
ring finger protein 149 |
| chr9_+_121567057 | 0.33 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr4_+_94974984 | 0.32 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr18_-_55635948 | 0.31 |
ENST00000565124.4
ENST00000398339.5 |
TCF4
|
transcription factor 4 |
| chr5_+_140821598 | 0.30 |
ENST00000614258.1
ENST00000529859.2 ENST00000529619.5 |
PCDHA5
|
protocadherin alpha 5 |
| chr1_+_207053229 | 0.30 |
ENST00000367080.8
ENST00000367079.3 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr6_+_121437378 | 0.30 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr2_+_90038848 | 0.30 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr20_-_31390580 | 0.29 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chr12_-_86256267 | 0.29 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr11_+_55827219 | 0.29 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor family 5 subfamily L member 2 |
| chr1_+_152811971 | 0.29 |
ENST00000360090.4
|
LCE1B
|
late cornified envelope 1B |
| chr12_+_26195313 | 0.28 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr5_-_35938572 | 0.27 |
ENST00000651391.1
ENST00000397366.5 ENST00000513623.5 ENST00000514524.2 ENST00000397367.6 |
CAPSL
|
calcyphosine like |
| chr11_+_55811367 | 0.27 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr4_-_159035226 | 0.27 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr14_+_22202561 | 0.27 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr17_+_39628496 | 0.27 |
ENST00000394265.5
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr3_+_42979281 | 0.27 |
ENST00000488863.5
ENST00000430121.3 |
GASK1A
|
golgi associated kinase 1A |
| chr14_+_22508602 | 0.27 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr11_-_129024157 | 0.26 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr2_+_90172802 | 0.26 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr5_+_151259793 | 0.26 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr20_-_51802433 | 0.25 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr5_+_55853314 | 0.25 |
ENST00000354961.8
ENST00000297015.7 |
IL31RA
|
interleukin 31 receptor A |
| chr6_+_26402237 | 0.24 |
ENST00000476549.6
ENST00000450085.6 ENST00000425234.6 ENST00000427334.5 ENST00000506698.1 ENST00000289361.11 |
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr12_+_6904962 | 0.24 |
ENST00000415834.5
ENST00000436789.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr20_-_51802509 | 0.24 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chrX_+_115593570 | 0.23 |
ENST00000539310.5
|
PLS3
|
plastin 3 |
| chr2_+_90234809 | 0.23 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr19_-_35812838 | 0.23 |
ENST00000653904.2
|
PRODH2
|
proline dehydrogenase 2 |
| chr8_-_7430348 | 0.22 |
ENST00000318124.3
|
DEFB103B
|
defensin beta 103B |
| chr13_+_48037692 | 0.21 |
ENST00000258662.3
|
NUDT15
|
nudix hydrolase 15 |
| chr12_-_21910853 | 0.21 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr3_+_119579676 | 0.21 |
ENST00000357003.7
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr11_-_16356538 | 0.20 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr2_+_102418642 | 0.20 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr2_+_102337148 | 0.19 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr16_-_28597042 | 0.19 |
ENST00000533150.5
ENST00000335715.9 |
SULT1A2
|
sulfotransferase family 1A member 2 |
| chr3_+_122055355 | 0.19 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr10_+_125896549 | 0.18 |
ENST00000368693.6
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr9_+_72577939 | 0.18 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr18_+_58341038 | 0.18 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr15_+_22015233 | 0.18 |
ENST00000639059.1
ENST00000640156.1 |
ENSG00000285472.1
ENSG00000284500.3
|
novel protein novel transcript |
| chr17_-_66229380 | 0.18 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr17_+_18183803 | 0.17 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr6_+_26183750 | 0.17 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr12_+_26195543 | 0.17 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr17_-_5035418 | 0.16 |
ENST00000254853.10
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 member 1 |
| chr7_-_88226965 | 0.16 |
ENST00000490437.5
ENST00000431660.5 |
SRI
|
sorcin |
| chr5_+_141177790 | 0.16 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr6_+_127577168 | 0.16 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr7_-_87713287 | 0.15 |
ENST00000416177.1
ENST00000265724.8 ENST00000543898.5 |
ABCB1
|
ATP binding cassette subfamily B member 1 |
| chr7_-_88226987 | 0.15 |
ENST00000394641.7
|
SRI
|
sorcin |
| chr12_+_12357418 | 0.15 |
ENST00000298571.6
|
BORCS5
|
BLOC-1 related complex subunit 5 |
| chr1_-_92486916 | 0.14 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr7_-_44541262 | 0.14 |
ENST00000289547.8
ENST00000546276.5 ENST00000423141.1 |
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr6_+_26402289 | 0.14 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr9_-_112332962 | 0.14 |
ENST00000458258.5
ENST00000210227.4 |
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr19_-_3557563 | 0.14 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_-_119322128 | 0.14 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr6_+_26087281 | 0.13 |
ENST00000353147.9
ENST00000397022.7 ENST00000352392.8 ENST00000349999.8 ENST00000317896.11 ENST00000470149.5 ENST00000336625.12 ENST00000461397.5 ENST00000488199.5 |
HFE
|
homeostatic iron regulator |
| chr15_+_21579912 | 0.13 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203 |
| chr19_-_14848922 | 0.13 |
ENST00000641129.1
|
OR7A10
|
olfactory receptor family 7 subfamily A member 10 |
| chr11_-_72112068 | 0.13 |
ENST00000537644.5
ENST00000538919.5 ENST00000539395.1 ENST00000542531.5 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr2_+_112542413 | 0.13 |
ENST00000417433.6
ENST00000263331.10 |
POLR1B
|
RNA polymerase I subunit B |
| chr7_+_107583919 | 0.13 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr1_+_207325629 | 0.13 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr2_+_87338511 | 0.13 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr7_+_118224654 | 0.13 |
ENST00000265224.9
ENST00000486422.1 ENST00000417525.5 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr12_+_26195647 | 0.13 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr12_-_9869345 | 0.13 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr6_+_37929959 | 0.12 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr2_-_88979016 | 0.12 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr10_+_113126670 | 0.12 |
ENST00000369389.6
|
TCF7L2
|
transcription factor 7 like 2 |
| chr22_-_39893755 | 0.12 |
ENST00000325157.7
|
ENTHD1
|
ENTH domain containing 1 |
| chr1_-_66801276 | 0.11 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr6_+_26087417 | 0.11 |
ENST00000357618.10
ENST00000309234.10 |
HFE
|
homeostatic iron regulator |
| chr1_-_230869564 | 0.11 |
ENST00000470540.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr19_+_55769118 | 0.11 |
ENST00000341750.5
|
RFPL4AL1
|
ret finger protein like 4A like 1 |
| chr11_-_107858777 | 0.11 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr6_+_154039333 | 0.11 |
ENST00000428397.6
|
OPRM1
|
opioid receptor mu 1 |
| chr1_-_207052980 | 0.11 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr17_-_40665121 | 0.11 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr17_+_18183052 | 0.11 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr12_-_118359639 | 0.11 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr1_-_23799533 | 0.11 |
ENST00000429356.5
|
GALE
|
UDP-galactose-4-epimerase |
| chr8_-_673547 | 0.10 |
ENST00000522893.1
|
ERICH1
|
glutamate rich 1 |
| chr4_-_46124046 | 0.10 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1 |
| chr16_+_31259922 | 0.10 |
ENST00000648685.1
ENST00000544665.9 |
ITGAM
|
integrin subunit alpha M |
| chr14_+_32329341 | 0.10 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr4_+_87799546 | 0.10 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr15_+_58410543 | 0.10 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr11_+_35180279 | 0.10 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_72112669 | 0.09 |
ENST00000545944.5
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr1_-_201171545 | 0.09 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr10_-_49188380 | 0.09 |
ENST00000374153.7
ENST00000374148.1 ENST00000374151.7 |
TMEM273
|
transmembrane protein 273 |
| chr12_-_8066331 | 0.09 |
ENST00000546241.1
ENST00000307637.5 |
C3AR1
|
complement C3a receptor 1 |
| chr12_+_6904733 | 0.09 |
ENST00000007969.12
ENST00000622489.4 ENST00000443597.7 ENST00000323702.9 |
LRRC23
|
leucine rich repeat containing 23 |
| chr3_-_101320558 | 0.09 |
ENST00000193391.8
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr19_-_40413364 | 0.09 |
ENST00000291825.11
ENST00000324001.8 |
PRX
|
periaxin |
| chr3_-_20012250 | 0.09 |
ENST00000389050.5
|
PP2D1
|
protein phosphatase 2C like domain containing 1 |
| chr6_-_110179995 | 0.09 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr3_-_64019334 | 0.09 |
ENST00000480205.5
|
PSMD6
|
proteasome 26S subunit, non-ATPase 6 |
| chr3_+_51817596 | 0.09 |
ENST00000456080.5
|
ENSG00000285749.2
|
novel protein |
| chr11_+_57712574 | 0.09 |
ENST00000278422.9
ENST00000378312.8 |
TMX2
|
thioredoxin related transmembrane protein 2 |
| chr12_+_28257195 | 0.09 |
ENST00000381259.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr2_-_169031317 | 0.09 |
ENST00000650372.1
|
ABCB11
|
ATP binding cassette subfamily B member 11 |
| chr9_-_28670285 | 0.09 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr16_-_81220370 | 0.09 |
ENST00000337114.8
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chr8_+_49911604 | 0.08 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr19_+_42076129 | 0.08 |
ENST00000359044.5
|
ZNF574
|
zinc finger protein 574 |
| chr11_-_72112750 | 0.08 |
ENST00000545680.5
ENST00000543587.5 ENST00000538393.5 ENST00000535234.5 ENST00000227618.8 ENST00000535503.5 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr2_-_219399981 | 0.08 |
ENST00000519905.1
ENST00000523282.5 ENST00000434339.5 ENST00000457935.5 |
DNPEP
|
aspartyl aminopeptidase |
| chr7_-_44541318 | 0.08 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr5_-_95081482 | 0.08 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr8_+_49911396 | 0.08 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr1_-_23799561 | 0.08 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr2_+_186506713 | 0.08 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr12_-_23584600 | 0.08 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr12_-_122500520 | 0.08 |
ENST00000540586.1
ENST00000543897.5 |
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr11_-_124315099 | 0.08 |
ENST00000641897.1
|
OR8D1
|
olfactory receptor family 8 subfamily D member 1 |
| chr17_-_59151794 | 0.07 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr16_+_15395745 | 0.07 |
ENST00000287594.7
ENST00000396385.4 ENST00000568766.1 |
MPV17L
ENSG00000261130.5
|
MPV17 mitochondrial inner membrane protein like novel protein |
| chrX_-_152709260 | 0.07 |
ENST00000535353.3
ENST00000638741.1 ENST00000640702.1 |
CSAG2
|
CSAG family member 2 |
| chr2_+_233917371 | 0.07 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr1_-_150765785 | 0.07 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr3_+_195720867 | 0.07 |
ENST00000436408.6
|
MUC20
|
mucin 20, cell surface associated |
| chr12_+_16347102 | 0.07 |
ENST00000536371.5
ENST00000010404.6 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr11_+_112025968 | 0.07 |
ENST00000680411.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr14_+_39267377 | 0.07 |
ENST00000556148.5
ENST00000348007.7 |
MIA2
|
MIA SH3 domain ER export factor 2 |
| chrX_-_72239022 | 0.07 |
ENST00000373657.2
ENST00000334463.4 |
ERCC6L
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr11_+_102047422 | 0.06 |
ENST00000434758.7
ENST00000526781.5 ENST00000534360.1 |
CFAP300
|
cilia and flagella associated protein 300 |
| chr1_+_151762899 | 0.06 |
ENST00000635322.1
ENST00000321531.10 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr2_-_89085787 | 0.06 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr6_+_110180418 | 0.06 |
ENST00000368930.5
ENST00000307731.2 |
CDC40
|
cell division cycle 40 |
| chr2_+_165239388 | 0.06 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr12_+_8513499 | 0.06 |
ENST00000299665.3
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chr19_-_9107475 | 0.06 |
ENST00000641081.1
|
OR7G2
|
olfactory receptor family 7 subfamily G member 2 |
| chr11_-_102843597 | 0.06 |
ENST00000299855.10
|
MMP3
|
matrix metallopeptidase 3 |
| chr17_+_58169401 | 0.06 |
ENST00000641866.1
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr17_+_29941605 | 0.05 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr3_-_57226344 | 0.05 |
ENST00000495160.2
|
HESX1
|
HESX homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.2 | 3.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.7 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.5 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.4 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.2 | GO:1904440 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 2.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:0032761 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 1.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.0 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0051918 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 1.7 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 2.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.3 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.2 | 0.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.6 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 1.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.3 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.2 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |