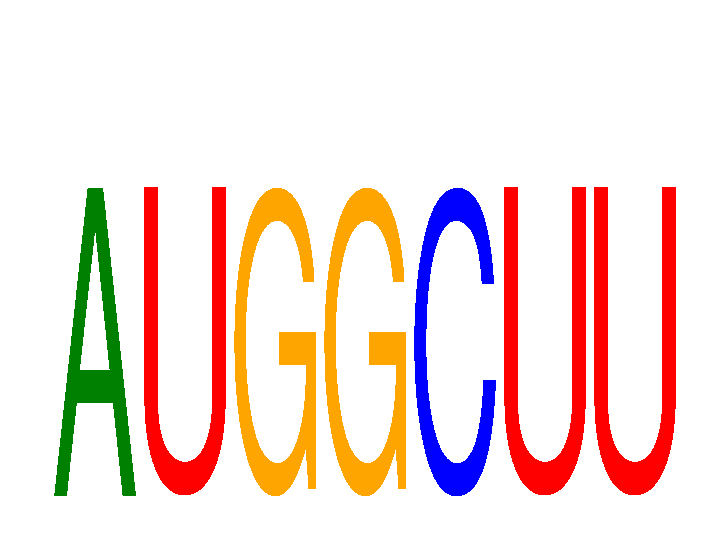Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for AUGGCUU
Z-value: 0.50
miRNA associated with seed AUGGCUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-135a-5p
|
MIMAT0000428 |
|
hsa-miR-135b-5p
|
MIMAT0000758 |
Activity profile of AUGGCUU motif
Sorted Z-values of AUGGCUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_2621766 | 0.26 |
ENST00000382100.8
|
VLDLR
|
very low density lipoprotein receptor |
| chr8_+_24914942 | 0.20 |
ENST00000433454.3
|
NEFM
|
neurofilament medium |
| chr19_+_3185911 | 0.20 |
ENST00000246117.9
ENST00000588428.5 |
NCLN
|
nicalin |
| chrX_+_153687918 | 0.19 |
ENST00000253122.10
|
SLC6A8
|
solute carrier family 6 member 8 |
| chr16_+_527698 | 0.19 |
ENST00000219611.7
ENST00000562370.5 ENST00000568988.5 |
CAPN15
|
calpain 15 |
| chr7_-_41703062 | 0.18 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr2_-_40452046 | 0.18 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr22_+_39994926 | 0.18 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F |
| chr17_-_58417521 | 0.17 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr6_-_99349647 | 0.17 |
ENST00000389677.6
|
FAXC
|
failed axon connections homolog, metaxin like GST domain containing |
| chr2_+_17878637 | 0.16 |
ENST00000304101.9
|
KCNS3
|
potassium voltage-gated channel modifier subfamily S member 3 |
| chr22_+_28883564 | 0.16 |
ENST00000544604.7
|
ZNRF3
|
zinc and ring finger 3 |
| chr7_-_84194781 | 0.16 |
ENST00000265362.9
|
SEMA3A
|
semaphorin 3A |
| chr4_-_110198650 | 0.15 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr8_+_103819244 | 0.15 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_-_55443263 | 0.15 |
ENST00000416792.2
ENST00000376325.10 |
SHISA7
|
shisa family member 7 |
| chr7_+_98106852 | 0.15 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr12_+_100794769 | 0.14 |
ENST00000392977.8
ENST00000546991.1 ENST00000392979.7 |
ANO4
|
anoctamin 4 |
| chr12_+_104456962 | 0.14 |
ENST00000547956.1
ENST00000549260.5 |
CHST11
|
carbohydrate sulfotransferase 11 |
| chr11_-_115504389 | 0.13 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr10_+_5412542 | 0.13 |
ENST00000355029.9
|
NET1
|
neuroepithelial cell transforming 1 |
| chrX_-_24672654 | 0.12 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr9_-_123184233 | 0.12 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr11_-_119729158 | 0.12 |
ENST00000264025.8
|
NECTIN1
|
nectin cell adhesion molecule 1 |
| chr22_+_49853801 | 0.12 |
ENST00000216268.6
|
ZBED4
|
zinc finger BED-type containing 4 |
| chr15_-_34336749 | 0.12 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr12_-_24949026 | 0.12 |
ENST00000539780.5
ENST00000546285.1 ENST00000342945.9 ENST00000261192.12 |
BCAT1
|
branched chain amino acid transaminase 1 |
| chr8_-_103415085 | 0.12 |
ENST00000297578.9
|
SLC25A32
|
solute carrier family 25 member 32 |
| chr3_+_3799424 | 0.11 |
ENST00000319331.4
|
LRRN1
|
leucine rich repeat neuronal 1 |
| chr6_-_116060859 | 0.11 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr13_-_75482151 | 0.11 |
ENST00000377636.8
|
TBC1D4
|
TBC1 domain family member 4 |
| chr1_-_93909329 | 0.11 |
ENST00000370238.8
ENST00000615724.1 |
GCLM
|
glutamate-cysteine ligase modifier subunit |
| chr10_+_95755652 | 0.10 |
ENST00000371207.8
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr8_-_118111806 | 0.10 |
ENST00000378204.7
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr8_+_97644164 | 0.10 |
ENST00000336273.8
|
MTDH
|
metadherin |
| chr17_-_46818680 | 0.10 |
ENST00000225512.6
|
WNT3
|
Wnt family member 3 |
| chr13_+_31199959 | 0.10 |
ENST00000343307.5
|
B3GLCT
|
beta 3-glucosyltransferase |
| chr5_+_149730260 | 0.10 |
ENST00000360453.8
ENST00000394320.7 ENST00000309241.10 |
PPARGC1B
|
PPARG coactivator 1 beta |
| chr8_-_20303955 | 0.10 |
ENST00000381569.5
|
LZTS1
|
leucine zipper tumor suppressor 1 |
| chr15_+_41774459 | 0.10 |
ENST00000457542.7
ENST00000456763.6 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr10_-_87818153 | 0.10 |
ENST00000308448.11
ENST00000680024.1 |
ATAD1
|
ATPase family AAA domain containing 1 |
| chr8_-_65842051 | 0.09 |
ENST00000401827.8
|
PDE7A
|
phosphodiesterase 7A |
| chr7_-_112790372 | 0.09 |
ENST00000449743.1
ENST00000441474.1 ENST00000312814.11 ENST00000454074.5 ENST00000447395.5 |
TMEM168
|
transmembrane protein 168 |
| chr3_-_79019444 | 0.09 |
ENST00000618833.4
ENST00000436010.6 ENST00000618846.4 |
ROBO1
|
roundabout guidance receptor 1 |
| chr2_-_47570905 | 0.09 |
ENST00000327876.5
|
KCNK12
|
potassium two pore domain channel subfamily K member 12 |
| chr8_+_93916882 | 0.09 |
ENST00000297598.5
ENST00000520728.5 ENST00000518107.5 ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_+_203626775 | 0.09 |
ENST00000367218.7
|
ATP2B4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr17_-_61928005 | 0.08 |
ENST00000444766.7
|
INTS2
|
integrator complex subunit 2 |
| chr7_-_140176970 | 0.08 |
ENST00000397560.7
|
KDM7A
|
lysine demethylase 7A |
| chr10_+_99659430 | 0.08 |
ENST00000370489.5
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr1_+_15617415 | 0.08 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2 |
| chr1_-_68497030 | 0.08 |
ENST00000456315.7
ENST00000525124.1 ENST00000370966.9 |
DEPDC1
|
DEP domain containing 1 |
| chr7_-_138981307 | 0.08 |
ENST00000440172.5
ENST00000422774.2 |
KIAA1549
|
KIAA1549 |
| chr2_-_128318860 | 0.08 |
ENST00000259241.7
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr13_+_42048645 | 0.08 |
ENST00000337343.9
ENST00000261491.9 ENST00000611224.1 |
DGKH
|
diacylglycerol kinase eta |
| chr3_+_113947901 | 0.08 |
ENST00000330212.7
ENST00000498275.5 |
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23 |
| chr12_+_4273751 | 0.08 |
ENST00000675880.1
ENST00000261254.8 |
CCND2
|
cyclin D2 |
| chr10_-_50623897 | 0.08 |
ENST00000361781.7
ENST00000429490.5 ENST00000619438.4 |
SGMS1
|
sphingomyelin synthase 1 |
| chr3_-_171460368 | 0.07 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr17_-_68291116 | 0.07 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chr4_+_145098269 | 0.07 |
ENST00000502586.5
ENST00000296577.9 |
ABCE1
|
ATP binding cassette subfamily E member 1 |
| chr1_-_184754808 | 0.07 |
ENST00000318130.13
ENST00000367512.7 |
EDEM3
|
ER degradation enhancing alpha-mannosidase like protein 3 |
| chr6_-_11044275 | 0.07 |
ENST00000354666.4
|
ELOVL2
|
ELOVL fatty acid elongase 2 |
| chr1_-_150235943 | 0.07 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr16_-_84504612 | 0.07 |
ENST00000562447.5
ENST00000343629.11 ENST00000565765.1 |
MEAK7
|
MTOR associated protein, eak-7 homolog |
| chr6_-_17706852 | 0.07 |
ENST00000262077.3
|
NUP153
|
nucleoporin 153 |
| chr12_-_54385727 | 0.07 |
ENST00000551109.5
ENST00000546970.5 |
ZNF385A
|
zinc finger protein 385A |
| chr8_-_8893548 | 0.07 |
ENST00000276282.7
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr11_+_20599602 | 0.07 |
ENST00000525748.6
|
SLC6A5
|
solute carrier family 6 member 5 |
| chr1_+_64745089 | 0.07 |
ENST00000294428.7
ENST00000371072.8 |
RAVER2
|
ribonucleoprotein, PTB binding 2 |
| chr20_-_49713842 | 0.07 |
ENST00000371711.4
|
B4GALT5
|
beta-1,4-galactosyltransferase 5 |
| chr1_+_171485520 | 0.07 |
ENST00000647382.2
ENST00000392078.7 ENST00000367742.7 ENST00000338920.8 |
PRRC2C
|
proline rich coiled-coil 2C |
| chr9_+_74497308 | 0.07 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chrX_+_12138426 | 0.07 |
ENST00000380682.5
ENST00000675598.1 |
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr17_-_7294592 | 0.07 |
ENST00000007699.10
|
YBX2
|
Y-box binding protein 2 |
| chr6_+_11093753 | 0.06 |
ENST00000416247.4
|
SMIM13
|
small integral membrane protein 13 |
| chr1_+_26826682 | 0.06 |
ENST00000374142.9
|
ZDHHC18
|
zinc finger DHHC-type palmitoyltransferase 18 |
| chr11_-_101583985 | 0.06 |
ENST00000344327.8
|
TRPC6
|
transient receptor potential cation channel subfamily C member 6 |
| chr13_+_94601830 | 0.06 |
ENST00000376958.5
|
GPR180
|
G protein-coupled receptor 180 |
| chr10_+_86756580 | 0.06 |
ENST00000372037.8
|
BMPR1A
|
bone morphogenetic protein receptor type 1A |
| chr6_-_107115493 | 0.06 |
ENST00000369042.6
|
BEND3
|
BEN domain containing 3 |
| chr15_+_59438149 | 0.06 |
ENST00000288228.10
ENST00000559628.5 ENST00000557914.5 ENST00000560474.5 |
FAM81A
|
family with sequence similarity 81 member A |
| chr17_+_7439504 | 0.06 |
ENST00000575331.1
ENST00000293829.9 |
ENSG00000272884.1
FGF11
|
novel transcript fibroblast growth factor 11 |
| chr2_+_32165841 | 0.06 |
ENST00000357055.7
ENST00000435660.5 ENST00000440718.5 ENST00000379343.6 ENST00000282587.9 ENST00000406369.2 |
SLC30A6
|
solute carrier family 30 member 6 |
| chr3_+_122795039 | 0.06 |
ENST00000261038.6
|
SLC49A4
|
solute carrier family 49 member 4 |
| chr17_+_75516514 | 0.06 |
ENST00000333213.11
ENST00000545228.3 ENST00000680999.1 |
TSEN54
|
tRNA splicing endonuclease subunit 54 |
| chr4_-_39032343 | 0.05 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr6_-_119349754 | 0.05 |
ENST00000368468.4
|
MAN1A1
|
mannosidase alpha class 1A member 1 |
| chr7_+_77696423 | 0.05 |
ENST00000334955.13
|
RSBN1L
|
round spermatid basic protein 1 like |
| chr6_-_10415043 | 0.05 |
ENST00000379613.10
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr12_-_57111338 | 0.05 |
ENST00000538913.6
ENST00000537215.6 ENST00000300134.8 ENST00000454075.7 ENST00000640254.2 ENST00000553275.1 ENST00000553533.2 |
STAT6
|
signal transducer and activator of transcription 6 |
| chr7_-_72336995 | 0.05 |
ENST00000329008.9
|
CALN1
|
calneuron 1 |
| chr2_+_182716227 | 0.05 |
ENST00000680258.1
ENST00000680667.1 ENST00000264065.12 ENST00000616986.5 ENST00000679884.1 |
DNAJC10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr8_-_29350666 | 0.05 |
ENST00000240100.7
|
DUSP4
|
dual specificity phosphatase 4 |
| chr16_+_72008588 | 0.05 |
ENST00000572887.5
ENST00000219240.9 ENST00000574309.5 ENST00000576145.1 |
DHODH
|
dihydroorotate dehydrogenase (quinone) |
| chr1_-_10796636 | 0.05 |
ENST00000377022.8
ENST00000344008.5 |
CASZ1
|
castor zinc finger 1 |
| chr15_-_68229658 | 0.05 |
ENST00000565471.6
ENST00000637494.1 ENST00000636314.1 ENST00000637667.1 ENST00000564752.1 ENST00000566347.5 ENST00000249806.11 ENST00000562767.2 |
CLN6
ENSG00000260007.3
|
CLN6 transmembrane ER protein novel protein |
| chr3_-_120094436 | 0.05 |
ENST00000264235.13
ENST00000677034.1 |
GSK3B
|
glycogen synthase kinase 3 beta |
| chr1_-_211830748 | 0.05 |
ENST00000366997.9
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr7_-_35695120 | 0.05 |
ENST00000311350.8
ENST00000396081.5 |
HERPUD2
|
HERPUD family member 2 |
| chr2_+_237627564 | 0.05 |
ENST00000308482.14
|
LRRFIP1
|
LRR binding FLII interacting protein 1 |
| chr15_+_32615501 | 0.05 |
ENST00000361627.8
ENST00000567348.5 ENST00000563864.5 ENST00000543522.5 |
ARHGAP11A
|
Rho GTPase activating protein 11A |
| chr6_-_85642922 | 0.05 |
ENST00000616122.5
ENST00000678618.1 ENST00000678816.1 ENST00000676630.1 ENST00000678589.1 ENST00000678878.1 ENST00000676542.1 ENST00000355238.11 ENST00000678899.1 |
SYNCRIP
|
synaptotagmin binding cytoplasmic RNA interacting protein |
| chr20_-_62065834 | 0.05 |
ENST00000252996.9
|
TAF4
|
TATA-box binding protein associated factor 4 |
| chr19_-_55117627 | 0.05 |
ENST00000263433.8
|
PPP1R12C
|
protein phosphatase 1 regulatory subunit 12C |
| chr21_-_41926680 | 0.05 |
ENST00000329623.11
|
C2CD2
|
C2 calcium dependent domain containing 2 |
| chr9_-_90642791 | 0.05 |
ENST00000375765.5
ENST00000636786.1 |
DIRAS2
|
DIRAS family GTPase 2 |
| chr2_+_48314637 | 0.05 |
ENST00000413569.5
ENST00000340553.8 |
FOXN2
|
forkhead box N2 |
| chr1_+_107141022 | 0.04 |
ENST00000370067.5
ENST00000370068.6 |
NTNG1
|
netrin G1 |
| chr13_+_79481124 | 0.04 |
ENST00000612570.4
ENST00000218652.11 |
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr2_+_128091166 | 0.04 |
ENST00000259253.11
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr1_-_157138388 | 0.04 |
ENST00000368192.9
|
ETV3
|
ETS variant transcription factor 3 |
| chr19_+_4007714 | 0.04 |
ENST00000262971.3
|
PIAS4
|
protein inhibitor of activated STAT 4 |
| chr3_-_136196305 | 0.04 |
ENST00000473093.1
ENST00000309993.3 |
MSL2
|
MSL complex subunit 2 |
| chr6_-_149864300 | 0.04 |
ENST00000239367.7
|
LRP11
|
LDL receptor related protein 11 |
| chr3_+_142723999 | 0.04 |
ENST00000476941.6
ENST00000273482.10 |
TRPC1
|
transient receptor potential cation channel subfamily C member 1 |
| chr10_-_60389833 | 0.04 |
ENST00000280772.7
|
ANK3
|
ankyrin 3 |
| chr3_-_151461011 | 0.04 |
ENST00000282466.4
|
IGSF10
|
immunoglobulin superfamily member 10 |
| chr10_+_119207560 | 0.04 |
ENST00000392870.3
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr18_+_8717371 | 0.04 |
ENST00000359865.7
|
MTCL1
|
microtubule crosslinking factor 1 |
| chr7_-_107563892 | 0.04 |
ENST00000297135.9
ENST00000605888.1 ENST00000347053.8 |
COG5
|
component of oligomeric golgi complex 5 |
| chr7_-_76358982 | 0.04 |
ENST00000307630.5
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma |
| chr14_-_74713041 | 0.04 |
ENST00000356357.9
ENST00000555249.1 ENST00000681599.1 ENST00000556202.5 ENST00000681099.1 |
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr10_-_689613 | 0.04 |
ENST00000280886.12
ENST00000634311.1 |
DIP2C
|
disco interacting protein 2 homolog C |
| chr11_+_121452291 | 0.04 |
ENST00000260197.12
|
SORL1
|
sortilin related receptor 1 |
| chr4_+_127782270 | 0.04 |
ENST00000508549.5
ENST00000296464.9 |
HSPA4L
|
heat shock protein family A (Hsp70) member 4 like |
| chr11_+_19117123 | 0.04 |
ENST00000399351.7
ENST00000446113.7 |
ZDHHC13
|
zinc finger DHHC-type palmitoyltransferase 13 |
| chr9_+_99105098 | 0.04 |
ENST00000374990.6
ENST00000374994.9 ENST00000552516.5 |
TGFBR1
|
transforming growth factor beta receptor 1 |
| chr1_-_202808406 | 0.04 |
ENST00000650569.1
ENST00000367265.9 ENST00000649770.1 |
KDM5B
|
lysine demethylase 5B |
| chr4_-_826113 | 0.04 |
ENST00000304062.11
|
CPLX1
|
complexin 1 |
| chr8_+_98117285 | 0.04 |
ENST00000401707.7
ENST00000522319.5 |
POP1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr9_-_10612966 | 0.04 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr1_+_42380772 | 0.04 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr9_+_126613922 | 0.04 |
ENST00000526117.6
ENST00000373474.9 ENST00000355497.10 |
LMX1B
|
LIM homeobox transcription factor 1 beta |
| chr22_+_21417357 | 0.04 |
ENST00000407464.7
|
HIC2
|
HIC ZBTB transcriptional repressor 2 |
| chr5_-_32312913 | 0.04 |
ENST00000280285.9
ENST00000382142.8 ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr18_-_23586422 | 0.04 |
ENST00000269228.10
|
NPC1
|
NPC intracellular cholesterol transporter 1 |
| chr11_+_64306227 | 0.04 |
ENST00000405666.5
ENST00000468670.2 |
ESRRA
|
estrogen related receptor alpha |
| chr7_+_4682252 | 0.04 |
ENST00000328914.5
|
FOXK1
|
forkhead box K1 |
| chr12_-_57633136 | 0.04 |
ENST00000341156.9
ENST00000550764.5 ENST00000551220.1 |
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyltransferase 1 |
| chr17_-_29294141 | 0.04 |
ENST00000225388.9
|
NUFIP2
|
nuclear FMR1 interacting protein 2 |
| chr19_+_589873 | 0.04 |
ENST00000251287.3
|
HCN2
|
hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2 |
| chr12_+_53380639 | 0.04 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr2_+_107986515 | 0.03 |
ENST00000409059.5
ENST00000264047.3 |
SLC5A7
|
solute carrier family 5 member 7 |
| chr10_+_91923762 | 0.03 |
ENST00000265990.11
|
BTAF1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr19_-_344786 | 0.03 |
ENST00000264819.7
|
MIER2
|
MIER family member 2 |
| chr6_+_138161932 | 0.03 |
ENST00000251691.5
|
ARFGEF3
|
ARFGEF family member 3 |
| chr3_+_43286512 | 0.03 |
ENST00000454177.5
ENST00000429705.6 ENST00000296088.12 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr2_-_152717966 | 0.03 |
ENST00000410080.8
|
PRPF40A
|
pre-mRNA processing factor 40 homolog A |
| chr17_+_12665882 | 0.03 |
ENST00000425538.6
|
MYOCD
|
myocardin |
| chr11_+_120236635 | 0.03 |
ENST00000260264.8
|
POU2F3
|
POU class 2 homeobox 3 |
| chr2_-_173965356 | 0.03 |
ENST00000310015.12
|
SP3
|
Sp3 transcription factor |
| chr14_-_93788475 | 0.03 |
ENST00000393140.6
|
PRIMA1
|
proline rich membrane anchor 1 |
| chr6_+_33420201 | 0.03 |
ENST00000644458.1
ENST00000449372.7 ENST00000628646.2 ENST00000418600.7 |
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr3_+_197960200 | 0.03 |
ENST00000482695.5
ENST00000330198.8 ENST00000419117.5 ENST00000420910.6 ENST00000332636.5 |
LMLN
|
leishmanolysin like peptidase |
| chr17_-_50707855 | 0.03 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr12_-_29381141 | 0.03 |
ENST00000546839.5
ENST00000552155.5 ENST00000360150.9 ENST00000550353.5 ENST00000548441.1 ENST00000552132.5 |
ERGIC2
|
ERGIC and golgi 2 |
| chr19_-_43639788 | 0.03 |
ENST00000222374.3
|
CADM4
|
cell adhesion molecule 4 |
| chr13_-_109786567 | 0.03 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr14_+_102592611 | 0.03 |
ENST00000262241.7
|
RCOR1
|
REST corepressor 1 |
| chr7_-_37916807 | 0.03 |
ENST00000436072.7
|
SFRP4
|
secreted frizzled related protein 4 |
| chr1_+_19882374 | 0.03 |
ENST00000375120.4
|
OTUD3
|
OTU deubiquitinase 3 |
| chr6_+_10555787 | 0.03 |
ENST00000316170.9
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr1_-_243255170 | 0.03 |
ENST00000366542.6
|
CEP170
|
centrosomal protein 170 |
| chr12_+_55818033 | 0.03 |
ENST00000552672.5
ENST00000243045.10 ENST00000550836.1 |
ORMDL2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr17_+_40062956 | 0.03 |
ENST00000450525.7
|
THRA
|
thyroid hormone receptor alpha |
| chr20_-_9838831 | 0.03 |
ENST00000378423.5
|
PAK5
|
p21 (RAC1) activated kinase 5 |
| chr18_+_62187247 | 0.03 |
ENST00000644646.2
ENST00000256858.10 ENST00000398130.6 |
RELCH
|
RAB11 binding and LisH domain, coiled-coil and HEAT repeat containing |
| chr14_+_56118404 | 0.03 |
ENST00000267460.9
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr7_-_32299287 | 0.03 |
ENST00000396193.5
|
PDE1C
|
phosphodiesterase 1C |
| chr1_-_220272415 | 0.03 |
ENST00000358951.7
|
RAB3GAP2
|
RAB3 GTPase activating non-catalytic protein subunit 2 |
| chr10_-_86521737 | 0.03 |
ENST00000298767.10
|
WAPL
|
WAPL cohesin release factor |
| chr12_+_57591158 | 0.03 |
ENST00000422156.7
ENST00000354947.10 ENST00000540759.6 ENST00000551772.5 ENST00000550465.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr15_+_26867047 | 0.03 |
ENST00000335625.10
ENST00000555182.5 ENST00000400081.7 |
GABRA5
|
gamma-aminobutyric acid type A receptor subunit alpha5 |
| chr17_+_42682470 | 0.03 |
ENST00000264638.9
|
CNTNAP1
|
contactin associated protein 1 |
| chr21_+_36699100 | 0.03 |
ENST00000290399.11
|
SIM2
|
SIM bHLH transcription factor 2 |
| chr1_-_244864560 | 0.03 |
ENST00000444376.7
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U |
| chr8_+_109540075 | 0.03 |
ENST00000614147.1
ENST00000337573.10 |
EBAG9
|
estrogen receptor binding site associated antigen 9 |
| chr7_-_87059639 | 0.03 |
ENST00000450689.7
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr6_+_151494007 | 0.03 |
ENST00000239374.8
|
CCDC170
|
coiled-coil domain containing 170 |
| chr15_-_72117712 | 0.03 |
ENST00000444904.5
ENST00000564571.5 |
MYO9A
|
myosin IXA |
| chr4_+_38664189 | 0.03 |
ENST00000514033.1
ENST00000261438.10 |
KLF3
|
Kruppel like factor 3 |
| chr4_+_169620527 | 0.03 |
ENST00000360642.7
ENST00000512813.5 ENST00000513761.6 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr13_-_49792675 | 0.03 |
ENST00000261667.8
|
KPNA3
|
karyopherin subunit alpha 3 |
| chr8_-_23457618 | 0.03 |
ENST00000358689.9
ENST00000518718.1 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr4_-_148444674 | 0.03 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chrX_+_41334154 | 0.03 |
ENST00000441189.4
ENST00000644513.1 ENST00000644109.1 ENST00000646122.1 ENST00000644074.1 ENST00000644876.2 ENST00000399959.7 ENST00000646319.1 |
DDX3X
|
DEAD-box helicase 3 X-linked |
| chr11_+_68460712 | 0.03 |
ENST00000528635.5
ENST00000533127.5 ENST00000529907.5 ENST00000529344.5 ENST00000534534.5 ENST00000524845.5 ENST00000393800.7 ENST00000265637.8 ENST00000524904.5 ENST00000393801.7 ENST00000265636.9 ENST00000529710.5 |
PPP6R3
|
protein phosphatase 6 regulatory subunit 3 |
| chr2_+_240435652 | 0.03 |
ENST00000264039.7
|
GPC1
|
glypican 1 |
| chr11_-_107018462 | 0.03 |
ENST00000526355.7
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr3_+_30606574 | 0.03 |
ENST00000295754.10
ENST00000359013.4 |
TGFBR2
|
transforming growth factor beta receptor 2 |
| chr11_-_61161414 | 0.03 |
ENST00000301765.10
|
VPS37C
|
VPS37C subunit of ESCRT-I |
| chr14_-_54441325 | 0.03 |
ENST00000556113.1
ENST00000553660.5 ENST00000216416.9 ENST00000395573.8 ENST00000557690.5 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr20_-_62937936 | 0.02 |
ENST00000266070.8
ENST00000370371.8 |
DIDO1
|
death inducer-obliterator 1 |
| chr8_-_142614469 | 0.02 |
ENST00000356613.4
|
ARC
|
activity regulated cytoskeleton associated protein |
| chr5_+_109689915 | 0.02 |
ENST00000261483.5
|
MAN2A1
|
mannosidase alpha class 2A member 1 |
| chr18_-_31943026 | 0.02 |
ENST00000582539.5
ENST00000582513.5 ENST00000283351.10 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr15_-_48178144 | 0.02 |
ENST00000616409.4
ENST00000324324.12 ENST00000610570.4 |
MYEF2
|
myelin expression factor 2 |
| chr16_+_22008083 | 0.02 |
ENST00000542527.7
ENST00000569656.5 ENST00000562695.1 |
MOSMO
|
modulator of smoothened |
| chr16_-_23149378 | 0.02 |
ENST00000219689.12
|
USP31
|
ubiquitin specific peptidase 31 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AUGGCUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.0 | 0.2 | GO:0048880 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) sensory system development(GO:0048880) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.0 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) response to L-arginine(GO:1903576) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.0 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.0 | GO:2000077 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.0 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.0 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.3 | GO:0051184 | cofactor transporter activity(GO:0051184) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |