Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
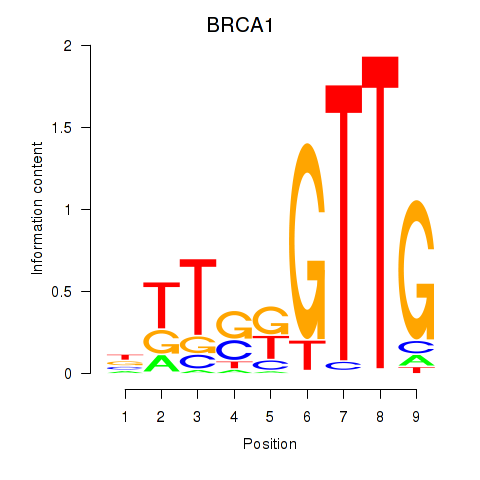
Results for BRCA1
Z-value: 0.49
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.23 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg38_v1_chr17_-_43125450_43125493 | -0.38 | 3.5e-01 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_159171607 | 1.12 |
ENST00000368124.8
ENST00000368125.9 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr14_-_59630806 | 0.53 |
ENST00000342503.8
|
RTN1
|
reticulon 1 |
| chr14_-_59630582 | 0.52 |
ENST00000395090.5
|
RTN1
|
reticulon 1 |
| chr9_-_92424427 | 0.45 |
ENST00000375550.5
|
OMD
|
osteomodulin |
| chr20_-_35438218 | 0.43 |
ENST00000374369.8
|
GDF5
|
growth differentiation factor 5 |
| chr11_-_27700472 | 0.32 |
ENST00000418212.5
ENST00000533246.5 |
BDNF
|
brain derived neurotrophic factor |
| chr11_-_27700447 | 0.32 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor |
| chr5_-_124746630 | 0.28 |
ENST00000513986.2
|
ZNF608
|
zinc finger protein 608 |
| chr7_-_23470469 | 0.24 |
ENST00000258729.8
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chrX_-_143635081 | 0.20 |
ENST00000338017.8
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chrX_+_10156960 | 0.19 |
ENST00000380833.9
|
CLCN4
|
chloride voltage-gated channel 4 |
| chr3_-_127736329 | 0.18 |
ENST00000398101.7
|
MGLL
|
monoglyceride lipase |
| chr7_+_100119607 | 0.16 |
ENST00000262932.5
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr2_-_144517663 | 0.16 |
ENST00000427902.5
ENST00000462355.2 ENST00000470879.5 ENST00000409487.7 ENST00000435831.5 ENST00000630572.2 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_151261839 | 0.15 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43 |
| chr21_+_44646414 | 0.15 |
ENST00000334670.9
|
KRTAP10-11
|
keratin associated protein 10-11 |
| chr14_+_88385643 | 0.14 |
ENST00000393545.9
ENST00000356583.9 ENST00000555401.5 ENST00000553885.5 |
SPATA7
|
spermatogenesis associated 7 |
| chr14_+_88385714 | 0.13 |
ENST00000045347.11
|
SPATA7
|
spermatogenesis associated 7 |
| chrX_-_71068384 | 0.13 |
ENST00000276105.3
ENST00000622259.4 |
SNX12
|
sorting nexin 12 |
| chr19_-_12801787 | 0.12 |
ENST00000334482.9
ENST00000301522.3 |
PRDX2
|
peroxiredoxin 2 |
| chr11_+_101914997 | 0.11 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chr20_+_58907981 | 0.10 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr5_+_154941063 | 0.10 |
ENST00000523037.6
ENST00000265229.12 ENST00000439747.7 ENST00000522038.5 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr12_-_80937918 | 0.10 |
ENST00000552864.6
|
LIN7A
|
lin-7 homolog A, crumbs cell polarity complex component |
| chr1_-_160262522 | 0.09 |
ENST00000440682.5
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr1_-_160262501 | 0.09 |
ENST00000447377.5
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chrX_-_71068311 | 0.09 |
ENST00000374274.8
|
SNX12
|
sorting nexin 12 |
| chr6_+_16129077 | 0.08 |
ENST00000356840.8
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr2_+_108378176 | 0.07 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr10_+_100535927 | 0.07 |
ENST00000299163.7
|
HIF1AN
|
hypoxia inducible factor 1 subunit alpha inhibitor |
| chr5_+_140700437 | 0.07 |
ENST00000274712.8
|
ZMAT2
|
zinc finger matrin-type 2 |
| chr19_-_37172432 | 0.07 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr11_+_73308237 | 0.07 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr12_-_122395422 | 0.07 |
ENST00000540304.6
|
CLIP1
|
CAP-Gly domain containing linker protein 1 |
| chr19_-_37172670 | 0.06 |
ENST00000588354.1
ENST00000292841.10 ENST00000356958.8 |
ZNF585A
|
zinc finger protein 585A |
| chr1_+_84181630 | 0.06 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_-_177196451 | 0.06 |
ENST00000430069.5
ENST00000630796.2 ENST00000428970.5 |
TBL1XR1
|
TBL1X receptor 1 |
| chr13_+_29428709 | 0.06 |
ENST00000542829.1
|
MTUS2
|
microtubule associated scaffold protein 2 |
| chr2_+_108377947 | 0.06 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr19_-_55580829 | 0.05 |
ENST00000592239.1
ENST00000325421.7 |
ZNF579
|
zinc finger protein 579 |
| chr6_-_43014254 | 0.05 |
ENST00000642748.1
|
MEA1
|
male-enhanced antigen 1 |
| chr11_+_33258304 | 0.05 |
ENST00000531504.5
ENST00000456517.2 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr19_-_47555856 | 0.05 |
ENST00000391901.7
ENST00000314121.8 |
ZNF541
|
zinc finger protein 541 |
| chr19_+_46347063 | 0.05 |
ENST00000012443.9
ENST00000391919.1 |
PPP5C
|
protein phosphatase 5 catalytic subunit |
| chr19_+_47130782 | 0.05 |
ENST00000597808.5
ENST00000413379.7 ENST00000600706.5 ENST00000270225.12 ENST00000598840.5 ENST00000600753.1 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr1_-_160262407 | 0.05 |
ENST00000419626.1
ENST00000610139.5 ENST00000475733.5 ENST00000407642.6 ENST00000368073.7 ENST00000326837.6 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr13_-_30464234 | 0.05 |
ENST00000399489.5
ENST00000339872.8 |
HMGB1
|
high mobility group box 1 |
| chr19_-_39834127 | 0.04 |
ENST00000601972.1
ENST00000430012.6 ENST00000323039.10 ENST00000348817.7 |
DYRK1B
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr17_+_38752731 | 0.04 |
ENST00000619426.5
ENST00000610434.4 |
PSMB3
|
proteasome 20S subunit beta 3 |
| chr18_-_26091175 | 0.04 |
ENST00000579061.5
ENST00000542420.6 |
SS18
|
SS18 subunit of BAF chromatin remodeling complex |
| chr14_+_22040576 | 0.04 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr20_-_49913693 | 0.04 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr7_-_100119323 | 0.04 |
ENST00000523306.5
ENST00000344095.8 ENST00000417349.5 ENST00000493322.5 ENST00000520135.5 ENST00000460673.2 ENST00000453269.7 ENST00000452041.5 ENST00000452438.6 ENST00000451699.5 |
TAF6
|
TATA-box binding protein associated factor 6 |
| chr13_-_49444004 | 0.04 |
ENST00000410043.5
ENST00000409308.6 |
CAB39L
|
calcium binding protein 39 like |
| chr2_+_135741814 | 0.04 |
ENST00000272638.14
|
UBXN4
|
UBX domain protein 4 |
| chr4_-_184649436 | 0.04 |
ENST00000308394.9
ENST00000517513.5 ENST00000447121.2 ENST00000393588.8 ENST00000523916.5 |
CASP3
|
caspase 3 |
| chr2_-_24972032 | 0.03 |
ENST00000534855.5
|
DNAJC27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr3_-_142448028 | 0.03 |
ENST00000392981.7
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr6_+_96563117 | 0.03 |
ENST00000450218.6
|
FHL5
|
four and a half LIM domains 5 |
| chr2_-_49974083 | 0.03 |
ENST00000636345.1
|
NRXN1
|
neurexin 1 |
| chr3_-_142448060 | 0.03 |
ENST00000264951.8
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr5_+_146203544 | 0.03 |
ENST00000506502.2
|
ENSG00000275740.1
|
novel readthrough transcript |
| chr11_-_72112750 | 0.03 |
ENST00000545680.5
ENST00000543587.5 ENST00000538393.5 ENST00000535234.5 ENST00000227618.8 ENST00000535503.5 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr1_-_160262540 | 0.02 |
ENST00000368074.6
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr10_-_24706622 | 0.02 |
ENST00000680286.1
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr4_+_109827963 | 0.02 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr16_-_740934 | 0.02 |
ENST00000540986.5
|
CIAO3
|
cytosolic iron-sulfur assembly component 3 |
| chr16_-_740980 | 0.02 |
ENST00000251588.7
|
CIAO3
|
cytosolic iron-sulfur assembly component 3 |
| chr10_+_120851341 | 0.02 |
ENST00000263461.11
|
WDR11
|
WD repeat domain 11 |
| chr2_-_24971900 | 0.02 |
ENST00000264711.7
|
DNAJC27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr6_-_158999748 | 0.02 |
ENST00000449822.5
|
RSPH3
|
radial spoke head 3 |
| chr10_+_90871946 | 0.01 |
ENST00000413330.5
ENST00000371703.8 ENST00000277882.7 |
RPP30
|
ribonuclease P/MRP subunit p30 |
| chr2_+_203014721 | 0.01 |
ENST00000683091.1
|
NBEAL1
|
neurobeachin like 1 |
| chr1_-_17045219 | 0.01 |
ENST00000491274.5
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr10_+_47322450 | 0.01 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr5_+_146203593 | 0.01 |
ENST00000265271.7
|
RBM27
|
RNA binding motif protein 27 |
| chr1_-_41242106 | 0.01 |
ENST00000337495.9
ENST00000372597.5 ENST00000372596.5 |
SCMH1
|
Scm polycomb group protein homolog 1 |
| chr1_+_171557845 | 0.01 |
ENST00000644916.1
|
PRRC2C
|
proline rich coiled-coil 2C |
| chr19_+_48469354 | 0.01 |
ENST00000452733.7
ENST00000641098.1 |
CYTH2
|
cytohesin 2 |
| chr9_-_83978429 | 0.01 |
ENST00000351839.7
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr2_+_203014842 | 0.00 |
ENST00000683969.1
ENST00000449802.5 |
NBEAL1
|
neurobeachin like 1 |
| chr14_+_20190896 | 0.00 |
ENST00000641879.2
ENST00000641682.1 |
OR11G2
|
olfactory receptor family 11 subfamily G member 2 |
| chr11_-_61430008 | 0.00 |
ENST00000394888.8
|
CPSF7
|
cleavage and polyadenylation specific factor 7 |
| chr10_-_100529854 | 0.00 |
ENST00000370320.4
ENST00000299166.9 ENST00000370322.5 |
NDUFB8
|
NADH:ubiquinone oxidoreductase subunit B8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.0 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |