Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for CDC5L
Z-value: 1.03
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.8 | cell division cycle 5 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg38_v1_chr6_+_44387686_44387730 | -0.61 | 1.1e-01 | Click! |
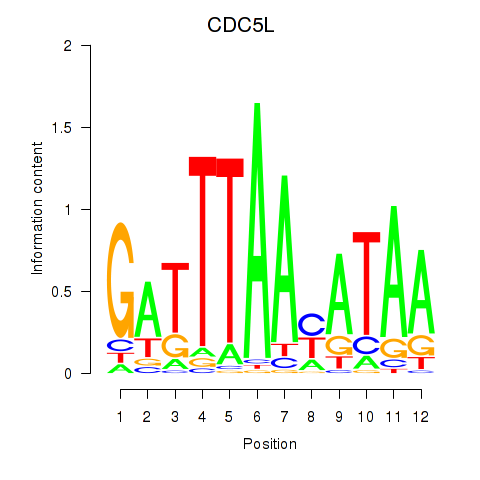
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_63887698 | 1.38 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr6_-_136526177 | 1.28 |
ENST00000617204.4
|
MAP7
|
microtubule associated protein 7 |
| chr4_-_15938740 | 1.20 |
ENST00000382333.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr6_-_136526472 | 1.18 |
ENST00000454590.5
ENST00000432797.6 |
MAP7
|
microtubule associated protein 7 |
| chr18_+_63775369 | 1.14 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr18_+_63775395 | 1.13 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr6_-_136525961 | 1.07 |
ENST00000438100.6
|
MAP7
|
microtubule associated protein 7 |
| chr2_-_112784486 | 1.06 |
ENST00000263339.4
|
IL1A
|
interleukin 1 alpha |
| chr18_+_63777773 | 0.99 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr17_-_7590072 | 0.96 |
ENST00000538513.6
ENST00000570788.1 ENST00000250055.3 |
SOX15
|
SRY-box transcription factor 15 |
| chr5_+_140786136 | 0.94 |
ENST00000378133.4
ENST00000504120.4 |
PCDHA1
|
protocadherin alpha 1 |
| chr5_+_167754918 | 0.92 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chr3_-_151316795 | 0.91 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr14_+_21990357 | 0.76 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr18_+_616672 | 0.71 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr3_+_122325237 | 0.69 |
ENST00000264474.4
ENST00000479204.1 |
CSTA
|
cystatin A |
| chr17_-_4739866 | 0.68 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chr5_+_140834230 | 0.68 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr10_-_114526897 | 0.67 |
ENST00000428430.1
ENST00000392952.7 |
ABLIM1
|
actin binding LIM protein 1 |
| chr18_+_616711 | 0.67 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr1_+_160400543 | 0.65 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr10_-_114684612 | 0.62 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr6_-_155455830 | 0.61 |
ENST00000159060.3
|
NOX3
|
NADPH oxidase 3 |
| chr10_-_114684457 | 0.60 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr8_+_32647080 | 0.56 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr21_+_42199686 | 0.56 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr8_+_117520832 | 0.55 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chrX_-_15664798 | 0.54 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator |
| chr1_-_161021096 | 0.54 |
ENST00000537746.1
ENST00000368026.11 |
F11R
|
F11 receptor |
| chr18_-_31102411 | 0.53 |
ENST00000251081.8
ENST00000280904.11 ENST00000682357.1 ENST00000648081.1 |
DSC2
|
desmocollin 2 |
| chr7_-_22194709 | 0.52 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr6_-_32816910 | 0.52 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr5_-_83673544 | 0.52 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_+_145936554 | 0.48 |
ENST00000359120.9
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr14_-_99271485 | 0.48 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr8_-_123737378 | 0.47 |
ENST00000419625.6
ENST00000262219.10 |
ANXA13
|
annexin A13 |
| chr8_-_15238423 | 0.47 |
ENST00000382080.6
|
SGCZ
|
sarcoglycan zeta |
| chr4_-_109801978 | 0.46 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr14_+_39233908 | 0.45 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr5_+_140848360 | 0.45 |
ENST00000532602.2
|
PCDHA9
|
protocadherin alpha 9 |
| chr8_-_85341705 | 0.44 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr7_+_18496269 | 0.44 |
ENST00000432645.6
|
HDAC9
|
histone deacetylase 9 |
| chr7_-_20217342 | 0.43 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr1_+_89364051 | 0.43 |
ENST00000370456.5
|
GBP6
|
guanylate binding protein family member 6 |
| chr8_+_18391276 | 0.42 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr8_+_103880412 | 0.40 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_-_53545091 | 0.39 |
ENST00000650454.1
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chrX_-_24672654 | 0.38 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr3_-_197260369 | 0.38 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr9_-_123184233 | 0.38 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr3_-_197573323 | 0.38 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr7_-_16881967 | 0.37 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr16_+_4795357 | 0.37 |
ENST00000586005.6
|
SMIM22
|
small integral membrane protein 22 |
| chr18_-_22417910 | 0.37 |
ENST00000391403.4
|
CTAGE1
|
cutaneous T cell lymphoma-associated antigen 1 |
| chr2_+_102736903 | 0.36 |
ENST00000639249.1
ENST00000454536.5 ENST00000409528.5 ENST00000409173.5 ENST00000488134.5 |
TMEM182
|
transmembrane protein 182 |
| chr4_-_121381007 | 0.36 |
ENST00000394427.3
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr8_+_103819244 | 0.35 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr15_-_22160868 | 0.35 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr6_-_131701401 | 0.35 |
ENST00000315453.4
|
OR2A4
|
olfactory receptor family 2 subfamily A member 4 |
| chr14_-_106737547 | 0.35 |
ENST00000632209.1
|
IGHV1-69-2
|
immunoglobulin heavy variable 1-69-2 |
| chr8_+_22053543 | 0.35 |
ENST00000519850.5
ENST00000381470.7 |
DMTN
|
dematin actin binding protein |
| chr2_+_230759918 | 0.34 |
ENST00000614925.1
|
CAB39
|
calcium binding protein 39 |
| chr4_-_99352730 | 0.34 |
ENST00000510055.5
ENST00000515683.6 ENST00000511397.3 |
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide |
| chr12_-_39340963 | 0.33 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A |
| chr6_+_27824084 | 0.33 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr6_+_132538290 | 0.33 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr1_+_244352627 | 0.33 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr1_-_226739271 | 0.32 |
ENST00000429204.6
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr4_+_68447453 | 0.32 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr14_-_21098848 | 0.32 |
ENST00000556174.5
ENST00000554478.5 ENST00000553980.1 ENST00000421093.6 |
ZNF219
|
zinc finger protein 219 |
| chr10_-_114526804 | 0.32 |
ENST00000369266.7
ENST00000369253.6 |
ABLIM1
|
actin binding LIM protein 1 |
| chr3_-_108953762 | 0.32 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr8_+_74824526 | 0.32 |
ENST00000649643.1
ENST00000260113.7 |
PI15
|
peptidase inhibitor 15 |
| chr5_+_55102635 | 0.32 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr7_-_144259722 | 0.31 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor family 2 subfamily A member 7 |
| chr3_-_197226351 | 0.31 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr4_-_142305935 | 0.31 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr6_+_41637005 | 0.31 |
ENST00000419164.6
ENST00000373051.6 |
MDFI
|
MyoD family inhibitor |
| chr11_-_102780620 | 0.31 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr5_+_140794832 | 0.30 |
ENST00000378132.2
ENST00000526136.2 ENST00000520672.2 |
PCDHA2
|
protocadherin alpha 2 |
| chr14_+_70452161 | 0.30 |
ENST00000603540.2
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr5_+_140786291 | 0.30 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1 |
| chr15_+_80072559 | 0.30 |
ENST00000560228.5
ENST00000559835.5 ENST00000559775.5 ENST00000558688.5 ENST00000560392.5 ENST00000560976.5 ENST00000558272.5 ENST00000558390.5 |
ZFAND6
|
zinc finger AN1-type containing 6 |
| chr1_+_92168915 | 0.29 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8 |
| chr11_-_13496018 | 0.29 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr18_+_58221535 | 0.29 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr18_-_36067524 | 0.29 |
ENST00000590898.5
ENST00000357384.8 ENST00000399022.9 ENST00000588737.5 |
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr4_-_149815826 | 0.29 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr3_-_119677346 | 0.29 |
ENST00000484810.5
ENST00000497116.1 ENST00000261070.7 |
COX17
|
cytochrome c oxidase copper chaperone COX17 |
| chr2_-_55917699 | 0.28 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr16_+_4795378 | 0.28 |
ENST00000588606.5
|
SMIM22
|
small integral membrane protein 22 |
| chr8_-_42501224 | 0.28 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr13_+_31739520 | 0.28 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr18_+_58341038 | 0.28 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr12_-_94650506 | 0.27 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr12_-_86838867 | 0.27 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C |
| chr12_-_123272234 | 0.27 |
ENST00000544658.5
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr13_+_31739542 | 0.26 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr19_-_32869741 | 0.26 |
ENST00000590341.5
ENST00000587772.1 ENST00000023064.9 |
SLC7A9
|
solute carrier family 7 member 9 |
| chr8_-_85341659 | 0.25 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr8_-_86230360 | 0.25 |
ENST00000419776.2
ENST00000297524.8 |
SLC7A13
|
solute carrier family 7 member 13 |
| chr2_-_75569711 | 0.25 |
ENST00000233712.5
|
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chr7_-_122699108 | 0.24 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr2_+_237968770 | 0.24 |
ENST00000434655.5
ENST00000612130.4 |
UBE2F
|
ubiquitin conjugating enzyme E2 F (putative) |
| chr11_-_85719111 | 0.24 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr6_-_32589833 | 0.24 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr5_+_140801028 | 0.24 |
ENST00000532566.3
ENST00000522353.3 |
PCDHA3
|
protocadherin alpha 3 |
| chr4_+_146175702 | 0.24 |
ENST00000296581.11
ENST00000649747.1 ENST00000502781.5 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr14_-_67412112 | 0.23 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr2_+_72887368 | 0.23 |
ENST00000234454.6
|
SPR
|
sepiapterin reductase |
| chr22_+_20774092 | 0.23 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr8_-_101204697 | 0.23 |
ENST00000517844.5
|
ZNF706
|
zinc finger protein 706 |
| chr6_+_36029082 | 0.23 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr16_-_71577082 | 0.22 |
ENST00000355962.5
|
TAT
|
tyrosine aminotransferase |
| chr8_-_109680812 | 0.22 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr3_+_66220984 | 0.22 |
ENST00000354883.11
ENST00000336733.10 |
SLC25A26
|
solute carrier family 25 member 26 |
| chr11_-_62841809 | 0.22 |
ENST00000525239.5
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr22_+_28772664 | 0.22 |
ENST00000448492.6
ENST00000421503.6 ENST00000249064.9 ENST00000444523.1 |
CCDC117
|
coiled-coil domain containing 117 |
| chr4_-_89029881 | 0.22 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A |
| chr3_+_35679690 | 0.21 |
ENST00000413378.5
ENST00000417925.5 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr12_-_49069970 | 0.21 |
ENST00000301068.11
|
RHEBL1
|
RHEB like 1 |
| chr7_-_16465728 | 0.21 |
ENST00000307068.5
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr11_-_101129706 | 0.21 |
ENST00000534013.5
|
PGR
|
progesterone receptor |
| chr4_+_41612892 | 0.21 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_181022842 | 0.20 |
ENST00000258301.6
|
STX6
|
syntaxin 6 |
| chr5_-_35938572 | 0.20 |
ENST00000651391.1
ENST00000397366.5 ENST00000513623.5 ENST00000514524.2 ENST00000397367.6 |
CAPSL
|
calcyphosine like |
| chr22_+_26483851 | 0.20 |
ENST00000215917.11
|
SRRD
|
SRR1 domain containing |
| chr9_-_19786928 | 0.20 |
ENST00000341998.6
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 member 2 |
| chr4_-_142305826 | 0.20 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_-_243843164 | 0.20 |
ENST00000491219.6
ENST00000680056.1 ENST00000492957.2 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr1_-_216805367 | 0.20 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr17_+_7884783 | 0.20 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chrX_+_139530730 | 0.20 |
ENST00000218099.7
|
F9
|
coagulation factor IX |
| chr21_-_30480364 | 0.19 |
ENST00000390689.3
|
KRTAP19-1
|
keratin associated protein 19-1 |
| chr1_+_99646025 | 0.19 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr4_+_145625992 | 0.19 |
ENST00000541599.5
|
MMAA
|
metabolism of cobalamin associated A |
| chr8_+_32646838 | 0.19 |
ENST00000651333.1
ENST00000652592.1 |
NRG1
|
neuregulin 1 |
| chr4_+_154563003 | 0.19 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr6_+_34889228 | 0.19 |
ENST00000360359.5
ENST00000649117.1 ENST00000650178.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr4_-_122456725 | 0.18 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chrX_-_132128020 | 0.18 |
ENST00000298542.9
|
FRMD7
|
FERM domain containing 7 |
| chr5_+_148268830 | 0.18 |
ENST00000511106.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chr3_-_98517096 | 0.18 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr9_+_132978651 | 0.18 |
ENST00000636137.1
|
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr11_-_114595777 | 0.17 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr11_+_60056653 | 0.17 |
ENST00000278865.8
|
MS4A3
|
membrane spanning 4-domains A3 |
| chr9_-_21385395 | 0.17 |
ENST00000380206.4
|
IFNA2
|
interferon alpha 2 |
| chr10_+_113129285 | 0.17 |
ENST00000637574.1
|
TCF7L2
|
transcription factor 7 like 2 |
| chr11_-_85719045 | 0.17 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr7_+_1688119 | 0.17 |
ENST00000424383.4
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr5_+_148268741 | 0.17 |
ENST00000398450.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chr4_-_47981535 | 0.17 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr14_+_22207502 | 0.17 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr3_-_194351290 | 0.17 |
ENST00000429275.1
ENST00000323830.4 |
CPN2
|
carboxypeptidase N subunit 2 |
| chr8_+_98944403 | 0.17 |
ENST00000457907.3
ENST00000523368.5 ENST00000297565.8 ENST00000435298.6 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr2_+_165239388 | 0.17 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr14_+_22052503 | 0.16 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr10_+_94938649 | 0.16 |
ENST00000461906.1
ENST00000260682.8 |
CYP2C9
|
cytochrome P450 family 2 subfamily C member 9 |
| chr4_+_41538143 | 0.16 |
ENST00000503057.6
ENST00000511496.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_+_109136707 | 0.16 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr11_-_7830840 | 0.16 |
ENST00000641167.1
|
OR5P3
|
olfactory receptor family 5 subfamily P member 3 |
| chr9_-_38424446 | 0.16 |
ENST00000377694.2
|
IGFBPL1
|
insulin like growth factor binding protein like 1 |
| chr22_+_38681941 | 0.15 |
ENST00000216034.6
|
TOMM22
|
translocase of outer mitochondrial membrane 22 |
| chr6_+_138404206 | 0.15 |
ENST00000607197.6
ENST00000367697.7 |
HEBP2
|
heme binding protein 2 |
| chr6_+_110874775 | 0.15 |
ENST00000675380.1
ENST00000368882.8 ENST00000368877.9 ENST00000368885.8 ENST00000672937.2 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr7_+_144000320 | 0.15 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr4_+_94974984 | 0.15 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr2_-_240140649 | 0.15 |
ENST00000319460.2
|
OTOS
|
otospiralin |
| chr12_+_20810698 | 0.15 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr4_-_121952014 | 0.15 |
ENST00000379645.8
|
TRPC3
|
transient receptor potential cation channel subfamily C member 3 |
| chr2_-_162152404 | 0.15 |
ENST00000375497.3
|
GCG
|
glucagon |
| chr15_+_41774459 | 0.15 |
ENST00000457542.7
ENST00000456763.6 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr2_+_195656734 | 0.15 |
ENST00000409086.7
|
SLC39A10
|
solute carrier family 39 member 10 |
| chr21_-_30166782 | 0.14 |
ENST00000286808.5
|
CLDN17
|
claudin 17 |
| chr1_-_186461089 | 0.14 |
ENST00000391997.3
|
PDC
|
phosducin |
| chr9_-_111330224 | 0.14 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr4_+_105710809 | 0.14 |
ENST00000360505.9
ENST00000510865.5 ENST00000509336.5 |
GSTCD
|
glutathione S-transferase C-terminal domain containing |
| chr4_+_69931066 | 0.14 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr12_+_100503352 | 0.14 |
ENST00000551379.5
ENST00000188403.7 |
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr9_+_132978687 | 0.14 |
ENST00000372122.4
ENST00000372123.5 |
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr14_+_22314435 | 0.14 |
ENST00000390467.3
|
TRAV40
|
T cell receptor alpha variable 40 |
| chr21_-_14210948 | 0.14 |
ENST00000681601.1
|
LIPI
|
lipase I |
| chr10_-_11532275 | 0.14 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr10_-_60733467 | 0.14 |
ENST00000373827.6
|
ANK3
|
ankyrin 3 |
| chr11_-_13495984 | 0.14 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr12_-_91004965 | 0.14 |
ENST00000261172.8
|
EPYC
|
epiphycan |
| chr1_+_25272439 | 0.14 |
ENST00000648012.1
|
RHD
|
Rh blood group D antigen |
| chr3_+_46370854 | 0.14 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr19_+_35448251 | 0.14 |
ENST00000599180.3
|
FFAR2
|
free fatty acid receptor 2 |
| chr11_-_114595750 | 0.14 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr11_+_60056587 | 0.13 |
ENST00000395032.6
ENST00000358152.6 |
MS4A3
|
membrane spanning 4-domains A3 |
| chr9_-_101383558 | 0.13 |
ENST00000674556.1
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase |
| chr9_-_28670285 | 0.13 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr12_-_10388976 | 0.13 |
ENST00000540818.5
|
KLRK1
|
killer cell lectin like receptor K1 |
| chr3_+_44874606 | 0.13 |
ENST00000296125.9
|
TGM4
|
transglutaminase 4 |
| chr5_+_70025247 | 0.13 |
ENST00000380751.9
ENST00000380750.8 ENST00000503931.5 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B |
| chr15_+_89776326 | 0.13 |
ENST00000341735.5
|
MESP2
|
mesoderm posterior bHLH transcription factor 2 |
| chr7_+_130207847 | 0.13 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr1_+_152686123 | 0.13 |
ENST00000368780.4
|
LCE2B
|
late cornified envelope 2B |
| chr14_+_22202561 | 0.13 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 1.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 0.7 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.2 | 0.6 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.2 | 0.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 0.5 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 1.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.4 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.3 | GO:1904732 | regulation of electron carrier activity(GO:1904732) |
| 0.1 | 1.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.3 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.2 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.8 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.8 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.3 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.3 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 3.9 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:2000182 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.0 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:1903233 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.0 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.6 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 2.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0097414 | glial cytoplasmic inclusion(GO:0097409) classical Lewy body(GO:0097414) Lewy neurite(GO:0097462) Lewy body corona(GO:1990038) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0000799 | nuclear condensin complex(GO:0000799) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.5 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.0 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.0 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0000832 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 5.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |