|
chr11_-_118252279
Show fit
|
0.76 |
ENST00000525386.5
ENST00000527472.1
ENST00000278949.9
|
MPZL3
|
myelin protein zero like 3
|
|
chr6_-_11778781
Show fit
|
0.76 |
ENST00000414691.8
ENST00000229583.9
|
ADTRP
|
androgen dependent TFPI regulating protein
|
|
chr11_-_120123026
Show fit
|
0.75 |
ENST00000533302.5
|
TRIM29
|
tripartite motif containing 29
|
|
chr20_+_59835853
Show fit
|
0.73 |
ENST00000492611.5
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr15_+_43692886
Show fit
|
0.71 |
ENST00000434505.5
ENST00000411750.5
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr15_+_43593054
Show fit
|
0.70 |
ENST00000453782.5
ENST00000300283.10
ENST00000437924.5
|
CKMT1B
|
creatine kinase, mitochondrial 1B
|
|
chr16_+_67199104
Show fit
|
0.69 |
ENST00000360833.6
ENST00000652269.1
ENST00000393997.8
|
ELMO3
|
engulfment and cell motility 3
|
|
chr16_+_67199509
Show fit
|
0.67 |
ENST00000477898.5
|
ELMO3
|
engulfment and cell motility 3
|
|
chr12_-_122716790
Show fit
|
0.62 |
ENST00000528880.3
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr12_-_122703346
Show fit
|
0.53 |
ENST00000328880.6
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr2_-_160200310
Show fit
|
0.53 |
ENST00000620391.4
|
ITGB6
|
integrin subunit beta 6
|
|
chr2_-_160200251
Show fit
|
0.52 |
ENST00000428609.6
ENST00000409967.6
ENST00000283249.7
|
ITGB6
|
integrin subunit beta 6
|
|
chr14_+_61321571
Show fit
|
0.52 |
ENST00000332981.11
|
PRKCH
|
protein kinase C eta
|
|
chr10_-_96271508
Show fit
|
0.51 |
ENST00000427367.6
ENST00000413476.6
ENST00000371176.6
|
BLNK
|
B cell linker
|
|
chr3_+_170037982
Show fit
|
0.50 |
ENST00000355897.10
|
GPR160
|
G protein-coupled receptor 160
|
|
chr2_-_160200289
Show fit
|
0.49 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6
|
|
chr17_+_50532713
Show fit
|
0.48 |
ENST00000503690.5
ENST00000514874.5
ENST00000268933.8
|
EPN3
|
epsin 3
|
|
chr8_-_85341705
Show fit
|
0.46 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1
|
|
chr20_+_1894145
Show fit
|
0.44 |
ENST00000400068.7
|
SIRPA
|
signal regulatory protein alpha
|
|
chr2_-_46462
Show fit
|
0.41 |
ENST00000327669.5
|
FAM110C
|
family with sequence similarity 110 member C
|
|
chr1_-_161021096
Show fit
|
0.41 |
ENST00000537746.1
ENST00000368026.11
|
F11R
|
F11 receptor
|
|
chr20_+_1894462
Show fit
|
0.40 |
ENST00000622179.4
|
SIRPA
|
signal regulatory protein alpha
|
|
chr6_-_11779606
Show fit
|
0.39 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein
|
|
chr8_+_55879818
Show fit
|
0.38 |
ENST00000520220.6
ENST00000519728.6
|
LYN
|
LYN proto-oncogene, Src family tyrosine kinase
|
|
chr12_-_6631632
Show fit
|
0.38 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr21_+_42499600
Show fit
|
0.38 |
ENST00000398341.7
|
SLC37A1
|
solute carrier family 37 member 1
|
|
chr6_-_116060859
Show fit
|
0.38 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase
|
|
chr1_+_31410164
Show fit
|
0.36 |
ENST00000536859.5
|
SERINC2
|
serine incorporator 2
|
|
chr16_+_4795378
Show fit
|
0.35 |
ENST00000588606.5
|
SMIM22
|
small integral membrane protein 22
|
|
chr15_+_80695277
Show fit
|
0.35 |
ENST00000258884.5
ENST00000558464.1
|
ABHD17C
|
abhydrolase domain containing 17C, depalmitoylase
|
|
chr7_+_70596078
Show fit
|
0.35 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2
|
|
chr16_+_4795357
Show fit
|
0.34 |
ENST00000586005.6
|
SMIM22
|
small integral membrane protein 22
|
|
chr1_+_86424154
Show fit
|
0.33 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2
|
|
chr1_-_40665654
Show fit
|
0.32 |
ENST00000372684.8
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr19_+_7637099
Show fit
|
0.32 |
ENST00000595950.5
ENST00000221283.10
ENST00000441779.6
ENST00000414284.6
|
STXBP2
|
syntaxin binding protein 2
|
|
chr12_-_66678934
Show fit
|
0.32 |
ENST00000545666.5
ENST00000398016.7
ENST00000359742.9
ENST00000538211.5
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr10_-_96271553
Show fit
|
0.32 |
ENST00000224337.10
|
BLNK
|
B cell linker
|
|
chr19_+_6464229
Show fit
|
0.32 |
ENST00000600229.6
ENST00000356762.7
|
CRB3
|
crumbs cell polarity complex component 3
|
|
chr15_+_90201301
Show fit
|
0.30 |
ENST00000411539.6
|
SEMA4B
|
semaphorin 4B
|
|
chr1_+_205504644
Show fit
|
0.29 |
ENST00000429964.7
ENST00000443813.6
|
CDK18
|
cyclin dependent kinase 18
|
|
chr8_-_85341659
Show fit
|
0.28 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1
|
|
chr2_-_75560893
Show fit
|
0.28 |
ENST00000410113.5
ENST00000393913.8
|
EVA1A
|
eva-1 homolog A, regulator of programmed cell death
|
|
chr11_+_63986411
Show fit
|
0.28 |
ENST00000538426.6
ENST00000543004.5
|
OTUB1
|
OTU deubiquitinase, ubiquitin aldehyde binding 1
|
|
chr20_-_49915509
Show fit
|
0.27 |
ENST00000289431.10
|
SPATA2
|
spermatogenesis associated 2
|
|
chr19_+_35248994
Show fit
|
0.27 |
ENST00000427250.5
ENST00000605618.6
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr19_+_6464491
Show fit
|
0.27 |
ENST00000308243.7
|
CRB3
|
crumbs cell polarity complex component 3
|
|
chr19_+_35248879
Show fit
|
0.25 |
ENST00000347609.8
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr4_-_83334782
Show fit
|
0.25 |
ENST00000681769.1
ENST00000513463.1
ENST00000311412.10
|
HPSE
|
heparanase
|
|
chr12_-_50222694
Show fit
|
0.25 |
ENST00000552783.5
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr2_+_147844601
Show fit
|
0.25 |
ENST00000404590.1
|
ACVR2A
|
activin A receptor type 2A
|
|
chr12_-_50222348
Show fit
|
0.25 |
ENST00000552823.5
ENST00000552909.5
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr12_-_52949818
Show fit
|
0.24 |
ENST00000546897.5
ENST00000552551.5
|
KRT8
|
keratin 8
|
|
chr1_-_26960413
Show fit
|
0.24 |
ENST00000320567.6
|
KDF1
|
keratinocyte differentiation factor 1
|
|
chr1_+_94820341
Show fit
|
0.24 |
ENST00000446120.6
ENST00000271227.11
ENST00000527077.5
ENST00000529450.5
|
SLC44A3
|
solute carrier family 44 member 3
|
|
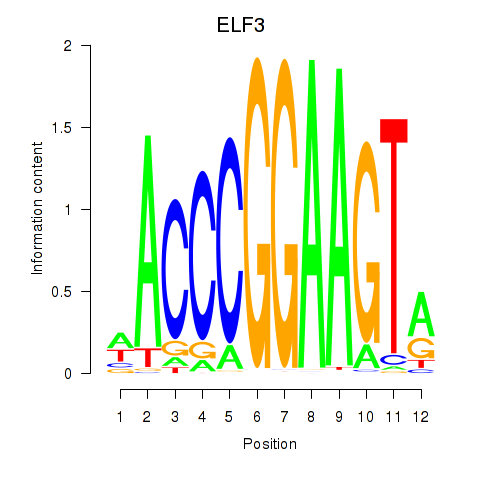
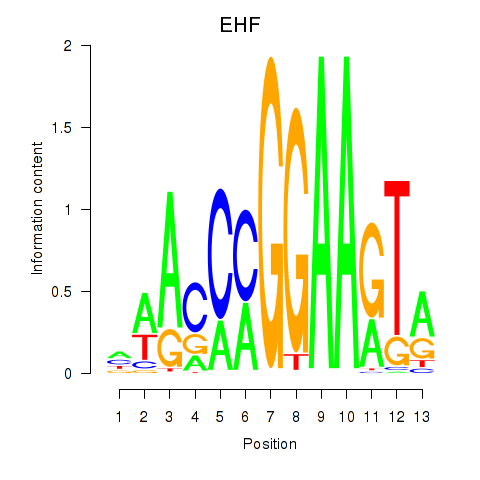
chr11_+_34621065
Show fit
|
0.24 |
ENST00000257831.8
|
EHF
|
ETS homologous factor
|
|
chr8_-_80080816
Show fit
|
0.23 |
ENST00000520527.5
ENST00000517427.5
ENST00000379097.7
ENST00000448733.3
|
TPD52
|
tumor protein D52
|
|
chr6_-_42451910
Show fit
|
0.23 |
ENST00000372922.8
ENST00000541110.5
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr15_+_45129933
Show fit
|
0.23 |
ENST00000321429.8
ENST00000389037.7
ENST00000558322.5
|
DUOX1
|
dual oxidase 1
|
|
chr1_-_26960369
Show fit
|
0.22 |
ENST00000616918.1
|
KDF1
|
keratinocyte differentiation factor 1
|
|
chr18_-_31162849
Show fit
|
0.22 |
ENST00000257197.7
ENST00000257198.6
|
DSC1
|
desmocollin 1
|
|
chr6_+_30880780
Show fit
|
0.22 |
ENST00000460944.6
ENST00000324771.12
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr5_+_40841308
Show fit
|
0.22 |
ENST00000381677.4
ENST00000254691.10
|
CARD6
|
caspase recruitment domain family member 6
|
|
chr2_+_168802610
Show fit
|
0.22 |
ENST00000397206.6
ENST00000317647.12
ENST00000397209.6
|
NOSTRIN
|
nitric oxide synthase trafficking
|
|
chr11_-_112164080
Show fit
|
0.22 |
ENST00000528832.1
ENST00000280357.12
|
IL18
|
interleukin 18
|
|
chr3_+_186996444
Show fit
|
0.22 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1
|
|
chr7_-_143408848
Show fit
|
0.22 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1
|
|
chr19_-_1174227
Show fit
|
0.21 |
ENST00000587024.5
ENST00000361757.8
|
SBNO2
|
strawberry notch homolog 2
|
|
chr6_-_143511682
Show fit
|
0.21 |
ENST00000002165.11
|
FUCA2
|
alpha-L-fucosidase 2
|
|
chr2_+_162344108
Show fit
|
0.21 |
ENST00000437150.7
ENST00000453113.6
|
GCA
|
grancalcin
|
|
chr11_-_112164056
Show fit
|
0.21 |
ENST00000524595.5
|
IL18
|
interleukin 18
|
|
chr15_+_74541200
Show fit
|
0.21 |
ENST00000622429.1
ENST00000346246.10
|
ARID3B
|
AT-rich interaction domain 3B
|
|
chr17_-_40100569
Show fit
|
0.21 |
ENST00000246672.4
|
NR1D1
|
nuclear receptor subfamily 1 group D member 1
|
|
chr1_+_17755307
Show fit
|
0.20 |
ENST00000375406.2
|
ACTL8
|
actin like 8
|
|
chr12_+_112907006
Show fit
|
0.20 |
ENST00000680455.1
ENST00000551241.6
ENST00000550689.2
ENST00000679841.1
ENST00000679494.1
ENST00000553185.2
|
OAS1
|
2'-5'-oligoadenylate synthetase 1
|
|
chr2_+_168802563
Show fit
|
0.20 |
ENST00000445023.6
|
NOSTRIN
|
nitric oxide synthase trafficking
|
|
chr11_-_7796942
Show fit
|
0.20 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2
|
|
chr3_+_113947901
Show fit
|
0.19 |
ENST00000330212.7
ENST00000498275.5
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23
|
|
chr2_+_147844488
Show fit
|
0.19 |
ENST00000535787.5
|
ACVR2A
|
activin A receptor type 2A
|
|
chr19_-_15934521
Show fit
|
0.19 |
ENST00000402119.9
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11
|
|
chr14_+_69398683
Show fit
|
0.19 |
ENST00000556605.5
ENST00000031146.8
ENST00000336643.10
|
SLC39A9
|
solute carrier family 39 member 9
|
|
chr19_-_15934410
Show fit
|
0.19 |
ENST00000326742.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11
|
|
chr6_+_109440695
Show fit
|
0.19 |
ENST00000258052.8
|
SMPD2
|
sphingomyelin phosphodiesterase 2
|
|
chr1_-_183590876
Show fit
|
0.19 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr10_-_102120246
Show fit
|
0.19 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1
|
|
chr5_+_150357629
Show fit
|
0.18 |
ENST00000650162.1
ENST00000377797.7
ENST00000445265.6
ENST00000323668.11
ENST00000643257.2
ENST00000646961.1
ENST00000513538.2
ENST00000439160.6
ENST00000394269.7
ENST00000427724.7
ENST00000504761.6
ENST00000513346.5
ENST00000515516.1
|
TCOF1
|
treacle ribosome biogenesis factor 1
|
|
chr11_+_35189869
Show fit
|
0.18 |
ENST00000525688.5
ENST00000278385.10
ENST00000533222.5
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr6_+_111259294
Show fit
|
0.18 |
ENST00000672303.1
ENST00000671876.2
ENST00000368847.5
|
MFSD4B
|
major facilitator superfamily domain containing 4B
|
|
chr1_-_153057504
Show fit
|
0.18 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A
|
|
chr16_+_57639518
Show fit
|
0.18 |
ENST00000540164.6
ENST00000568531.5
|
ADGRG1
|
adhesion G protein-coupled receptor G1
|
|
chr11_-_58578096
Show fit
|
0.18 |
ENST00000528954.5
ENST00000528489.1
|
LPXN
|
leupaxin
|
|
chr11_-_9314564
Show fit
|
0.18 |
ENST00000611268.4
ENST00000528080.6
ENST00000527813.5
ENST00000533723.1
|
TMEM41B
|
transmembrane protein 41B
|
|
chr17_-_4736380
Show fit
|
0.18 |
ENST00000576153.5
|
CXCL16
|
C-X-C motif chemokine ligand 16
|
|
chr3_+_113948004
Show fit
|
0.18 |
ENST00000638807.2
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23
|
|
chr1_+_23791138
Show fit
|
0.17 |
ENST00000374514.8
ENST00000420982.5
ENST00000374505.6
|
LYPLA2
|
lysophospholipase 2
|
|
chr11_-_72785932
Show fit
|
0.17 |
ENST00000539138.1
ENST00000542989.5
|
STARD10
|
StAR related lipid transfer domain containing 10
|
|
chr11_-_65117639
Show fit
|
0.17 |
ENST00000528598.1
ENST00000310597.6
|
ZNHIT2
|
zinc finger HIT-type containing 2
|
|
chr19_-_15934853
Show fit
|
0.17 |
ENST00000620614.4
ENST00000248041.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11
|
|
chr10_+_93566659
Show fit
|
0.17 |
ENST00000371481.9
ENST00000371483.8
ENST00000604414.1
|
FFAR4
|
free fatty acid receptor 4
|
|
chr16_-_19718175
Show fit
|
0.17 |
ENST00000219837.12
|
KNOP1
|
lysine rich nucleolar protein 1
|
|
chr1_+_43969970
Show fit
|
0.17 |
ENST00000255108.8
ENST00000396758.6
|
DPH2
|
diphthamide biosynthesis 2
|
|
chr8_-_133297092
Show fit
|
0.17 |
ENST00000522890.5
ENST00000675983.1
ENST00000518176.5
ENST00000323851.13
ENST00000522476.5
ENST00000518066.5
ENST00000521544.5
ENST00000674605.1
ENST00000518480.5
ENST00000523892.5
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr11_+_112176364
Show fit
|
0.17 |
ENST00000526088.5
ENST00000532593.5
ENST00000531169.5
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr19_-_55149193
Show fit
|
0.16 |
ENST00000587758.5
ENST00000588981.6
ENST00000356783.9
ENST00000291901.12
ENST00000588426.5
ENST00000536926.5
ENST00000588147.5
|
TNNT1
|
troponin T1, slow skeletal type
|
|
chr19_-_45730861
Show fit
|
0.16 |
ENST00000317683.4
|
FBXO46
|
F-box protein 46
|
|
chr2_+_37231798
Show fit
|
0.16 |
ENST00000439218.5
ENST00000432075.1
|
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7
|
|
chrX_+_139530730
Show fit
|
0.16 |
ENST00000218099.7
|
F9
|
coagulation factor IX
|
|
chr3_-_172523460
Show fit
|
0.16 |
ENST00000420541.6
|
TNFSF10
|
TNF superfamily member 10
|
|
chr3_+_66220984
Show fit
|
0.16 |
ENST00000354883.11
ENST00000336733.10
|
SLC25A26
|
solute carrier family 25 member 26
|
|
chr19_-_54173190
Show fit
|
0.16 |
ENST00000617472.4
|
TMC4
|
transmembrane channel like 4
|
|
chr18_+_58149314
Show fit
|
0.16 |
ENST00000435432.6
ENST00000357895.9
ENST00000586263.5
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr11_+_34621109
Show fit
|
0.16 |
ENST00000450654.6
|
EHF
|
ETS homologous factor
|
|
chr11_-_72781858
Show fit
|
0.16 |
ENST00000537947.5
|
STARD10
|
StAR related lipid transfer domain containing 10
|
|
chr12_+_4269771
Show fit
|
0.16 |
ENST00000676411.1
|
CCND2
|
cyclin D2
|
|
chr3_-_172523423
Show fit
|
0.16 |
ENST00000241261.7
|
TNFSF10
|
TNF superfamily member 10
|
|
chr2_-_237590694
Show fit
|
0.16 |
ENST00000264601.8
ENST00000411462.5
ENST00000409822.1
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr19_+_49335396
Show fit
|
0.16 |
ENST00000598095.5
ENST00000426897.6
ENST00000323906.9
ENST00000535669.6
ENST00000597602.1
ENST00000595660.1
|
CD37
|
CD37 molecule
|
|
chrX_+_83508284
Show fit
|
0.16 |
ENST00000644024.2
|
POU3F4
|
POU class 3 homeobox 4
|
|
chr21_+_42199686
Show fit
|
0.15 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1
|
|
chr6_-_109440504
Show fit
|
0.15 |
ENST00000520723.5
ENST00000518648.1
ENST00000417394.6
ENST00000521072.7
|
PPIL6
|
peptidylprolyl isomerase like 6
|
|
chr17_+_43483949
Show fit
|
0.15 |
ENST00000540306.5
ENST00000262415.8
|
DHX8
|
DEAH-box helicase 8
|
|
chr12_+_112906777
Show fit
|
0.15 |
ENST00000452357.7
ENST00000445409.7
|
OAS1
|
2'-5'-oligoadenylate synthetase 1
|
|
chr2_+_162344338
Show fit
|
0.15 |
ENST00000233612.8
|
GCA
|
grancalcin
|
|
chr1_-_209784521
Show fit
|
0.15 |
ENST00000294811.2
|
C1orf74
|
chromosome 1 open reading frame 74
|
|
chr14_+_24171853
Show fit
|
0.15 |
ENST00000620473.4
ENST00000557806.5
ENST00000611366.5
|
REC8
|
REC8 meiotic recombination protein
|
|
chr15_+_88635626
Show fit
|
0.15 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20
|
|
chr17_+_42562120
Show fit
|
0.15 |
ENST00000585811.1
ENST00000585909.1
ENST00000586771.1
ENST00000421097.6
ENST00000591779.5
ENST00000393818.3
ENST00000587858.5
ENST00000587214.1
ENST00000587157.1
ENST00000590958.5
|
COASY
|
Coenzyme A synthase
|
|
chr9_-_114387973
Show fit
|
0.15 |
ENST00000374088.8
|
AKNA
|
AT-hook transcription factor
|
|
chr3_-_79767987
Show fit
|
0.15 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1
|
|
chr5_+_69415065
Show fit
|
0.15 |
ENST00000647531.1
ENST00000645446.1
ENST00000325631.10
ENST00000454295.6
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr1_+_205504592
Show fit
|
0.15 |
ENST00000506784.5
ENST00000360066.6
|
CDK18
|
cyclin dependent kinase 18
|
|
chr11_+_67056805
Show fit
|
0.15 |
ENST00000308831.7
|
RHOD
|
ras homolog family member D
|
|
chr3_+_119498529
Show fit
|
0.15 |
ENST00000493694.1
ENST00000494664.6
|
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1
|
|
chr3_-_121660892
Show fit
|
0.15 |
ENST00000428394.6
ENST00000314583.8
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr19_+_58183029
Show fit
|
0.15 |
ENST00000424679.6
ENST00000617501.5
ENST00000345813.7
|
ZNF274
|
zinc finger protein 274
|
|
chr8_+_144477975
Show fit
|
0.15 |
ENST00000435887.2
|
PPP1R16A
|
protein phosphatase 1 regulatory subunit 16A
|
|
chr1_+_153778178
Show fit
|
0.14 |
ENST00000532853.5
|
SLC27A3
|
solute carrier family 27 member 3
|
|
chr3_-_47282752
Show fit
|
0.14 |
ENST00000456548.5
ENST00000432493.5
ENST00000684063.1
ENST00000444589.6
|
KIF9
|
kinesin family member 9
|
|
chr12_-_64222239
Show fit
|
0.14 |
ENST00000311915.12
ENST00000398055.8
ENST00000544871.1
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr18_-_70205655
Show fit
|
0.14 |
ENST00000255674.11
ENST00000640769.2
|
RTTN
|
rotatin
|
|
chr1_+_169367934
Show fit
|
0.14 |
ENST00000367807.7
ENST00000329281.6
ENST00000420531.1
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr16_-_352714
Show fit
|
0.14 |
ENST00000262320.8
|
AXIN1
|
axin 1
|
|
chr12_+_57591158
Show fit
|
0.14 |
ENST00000422156.7
ENST00000354947.10
ENST00000540759.6
ENST00000551772.5
ENST00000550465.5
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma
|
|
chr1_-_161045961
Show fit
|
0.14 |
ENST00000368021.7
|
USF1
|
upstream transcription factor 1
|
|
chr19_+_17281645
Show fit
|
0.14 |
ENST00000394458.7
ENST00000594072.6
|
ANKLE1
|
ankyrin repeat and LEM domain containing 1
|
|
chr17_-_7262343
Show fit
|
0.14 |
ENST00000571881.2
ENST00000360325.11
|
CLDN7
|
claudin 7
|
|
chr14_+_92513766
Show fit
|
0.14 |
ENST00000216487.12
ENST00000620541.4
ENST00000557762.1
|
RIN3
|
Ras and Rab interactor 3
|
|
chr6_+_47698574
Show fit
|
0.14 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4
|
|
chr4_+_15681815
Show fit
|
0.14 |
ENST00000422728.3
|
FAM200B
|
family with sequence similarity 200 member B
|
|
chr16_-_11587162
Show fit
|
0.14 |
ENST00000570904.5
ENST00000574701.5
|
LITAF
|
lipopolysaccharide induced TNF factor
|
|
chr6_+_47698538
Show fit
|
0.14 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4
|
|
chr6_-_166956561
Show fit
|
0.14 |
ENST00000366855.10
|
RNASET2
|
ribonuclease T2
|
|
chr5_-_83077343
Show fit
|
0.14 |
ENST00000502346.2
|
TMEM167A
|
transmembrane protein 167A
|
|
chr12_+_112906949
Show fit
|
0.13 |
ENST00000679971.1
ENST00000675868.2
ENST00000550883.2
ENST00000553152.2
ENST00000202917.10
ENST00000679467.1
ENST00000680659.1
ENST00000540589.3
ENST00000552526.2
ENST00000681228.1
ENST00000680934.1
ENST00000681700.1
ENST00000679987.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1
|
|
chr6_-_98947911
Show fit
|
0.13 |
ENST00000369244.7
ENST00000229971.2
|
FBXL4
|
F-box and leucine rich repeat protein 4
|
|
chr10_-_102120318
Show fit
|
0.13 |
ENST00000673968.1
|
LDB1
|
LIM domain binding 1
|
|
chr3_-_48430045
Show fit
|
0.13 |
ENST00000296440.11
|
PLXNB1
|
plexin B1
|
|
chr14_-_23567734
Show fit
|
0.13 |
ENST00000556843.1
ENST00000397120.8
ENST00000557189.5
|
AP1G2
|
adaptor related protein complex 1 subunit gamma 2
|
|
chr15_-_101652365
Show fit
|
0.13 |
ENST00000428002.6
ENST00000333202.8
ENST00000559107.5
ENST00000347970.7
|
TM2D3
|
TM2 domain containing 3
|
|
chr8_+_22053543
Show fit
|
0.13 |
ENST00000519850.5
ENST00000381470.7
|
DMTN
|
dematin actin binding protein
|
|
chr5_-_16508951
Show fit
|
0.13 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr4_-_102345469
Show fit
|
0.13 |
ENST00000356736.5
ENST00000682932.1
|
SLC39A8
|
solute carrier family 39 member 8
|
|
chr5_-_16508858
Show fit
|
0.13 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr11_-_72752376
Show fit
|
0.13 |
ENST00000393609.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr6_+_111259474
Show fit
|
0.13 |
ENST00000672554.1
ENST00000673024.1
|
MFSD4B
|
major facilitator superfamily domain containing 4B
|
|
chr7_+_75878993
Show fit
|
0.13 |
ENST00000318622.8
|
RHBDD2
|
rhomboid domain containing 2
|
|
chr19_-_50969567
Show fit
|
0.13 |
ENST00000310157.7
|
KLK6
|
kallikrein related peptidase 6
|
|
chr4_-_102345196
Show fit
|
0.13 |
ENST00000683412.1
ENST00000682227.1
|
SLC39A8
|
solute carrier family 39 member 8
|
|
chr5_-_16508812
Show fit
|
0.13 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr2_-_74440484
Show fit
|
0.13 |
ENST00000305557.9
ENST00000233330.6
|
RTKN
|
rhotekin
|
|
chr20_+_62238541
Show fit
|
0.12 |
ENST00000644775.1
ENST00000645442.1
ENST00000643412.1
ENST00000644702.1
|
OSBPL2
|
oxysterol binding protein like 2
|
|
chr2_+_105744876
Show fit
|
0.12 |
ENST00000233154.9
ENST00000451463.6
|
NCK2
|
NCK adaptor protein 2
|
|
chr10_+_127907036
Show fit
|
0.12 |
ENST00000254667.8
ENST00000442830.5
|
PTPRE
|
protein tyrosine phosphatase receptor type E
|
|
chr21_-_32612806
Show fit
|
0.12 |
ENST00000673807.1
|
CFAP298-TCP10L
|
CFAP298-TCP10L readthrough
|
|
chr11_+_45804038
Show fit
|
0.12 |
ENST00000442528.2
ENST00000526817.1
|
SLC35C1
|
solute carrier family 35 member C1
|
|
chr13_-_26222255
Show fit
|
0.12 |
ENST00000381588.9
|
RNF6
|
ring finger protein 6
|
|
chr16_+_14975061
Show fit
|
0.12 |
ENST00000455313.6
|
PDXDC1
|
pyridoxal dependent decarboxylase domain containing 1
|
|
chr17_+_7445055
Show fit
|
0.12 |
ENST00000306071.7
ENST00000572857.5
|
CHRNB1
|
cholinergic receptor nicotinic beta 1 subunit
|
|
chr16_+_82056423
Show fit
|
0.12 |
ENST00000568090.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2
|
|
chr17_-_40937445
Show fit
|
0.12 |
ENST00000436344.7
ENST00000485751.1
|
KRT23
|
keratin 23
|
|
chr5_-_16508990
Show fit
|
0.12 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1
|
|
chr10_+_73744346
Show fit
|
0.12 |
ENST00000345254.9
ENST00000339365.2
|
SEC24C
|
SEC24 homolog C, COPII coat complex component
|
|
chr10_-_117005570
Show fit
|
0.12 |
ENST00000260777.14
ENST00000392903.3
|
SHTN1
|
shootin 1
|
|
chr9_-_132406807
Show fit
|
0.12 |
ENST00000334270.3
ENST00000612514.4
|
TTF1
|
transcription termination factor 1
|
|
chr5_-_16508788
Show fit
|
0.12 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr3_+_119498575
Show fit
|
0.12 |
ENST00000466984.1
|
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1
|
|
chr19_-_51027662
Show fit
|
0.12 |
ENST00000594768.5
|
KLK11
|
kallikrein related peptidase 11
|
|
chr11_+_35189964
Show fit
|
0.11 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr7_+_73827737
Show fit
|
0.11 |
ENST00000435050.1
|
CLDN4
|
claudin 4
|
|
chr19_-_45178200
Show fit
|
0.11 |
ENST00000592647.1
ENST00000006275.8
ENST00000588062.5
ENST00000585934.1
|
TRAPPC6A
|
trafficking protein particle complex 6A
|
|
chr11_+_111918900
Show fit
|
0.11 |
ENST00000278601.6
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr7_+_75879009
Show fit
|
0.11 |
ENST00000006777.11
|
RHBDD2
|
rhomboid domain containing 2
|
|
chr8_-_98294195
Show fit
|
0.11 |
ENST00000430223.7
|
NIPAL2
|
NIPA like domain containing 2
|
|
chr17_+_4940118
Show fit
|
0.11 |
ENST00000572430.5
|
RNF167
|
ring finger protein 167
|
|
chr1_+_154974653
Show fit
|
0.11 |
ENST00000368439.5
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B
|
|
chr12_-_95116967
Show fit
|
0.11 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr2_+_105851748
Show fit
|
0.11 |
ENST00000425756.1
ENST00000393349.2
|
NCK2
|
NCK adaptor protein 2
|
|
chr3_+_53161120
Show fit
|
0.11 |
ENST00000394729.6
ENST00000330452.8
ENST00000652449.1
|
PRKCD
|
protein kinase C delta
|
|
chrX_+_80335504
Show fit
|
0.11 |
ENST00000538312.5
|
TENT5D
|
terminal nucleotidyltransferase 5D
|
|
chr17_-_40937641
Show fit
|
0.11 |
ENST00000209718.8
|
KRT23
|
keratin 23
|
|
chr17_+_41226648
Show fit
|
0.11 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2
|