Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for EVX2
Z-value: 0.50
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | even-skipped homeobox 2 |
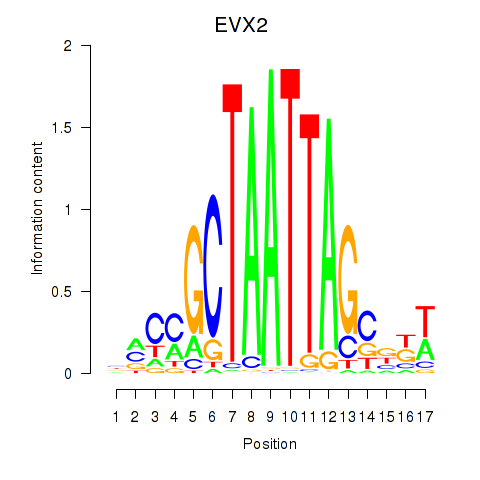
Activity profile of EVX2 motif
Sorted Z-values of EVX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_115504389 | 0.41 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr3_+_111999189 | 0.39 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr3_+_111998915 | 0.37 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr3_+_111998739 | 0.36 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr1_+_27934980 | 0.36 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr3_+_111999326 | 0.35 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr1_+_27935110 | 0.34 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr6_+_132538290 | 0.32 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr4_+_76435216 | 0.31 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr1_-_99766620 | 0.29 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr11_+_65787056 | 0.25 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr4_+_143433491 | 0.24 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr20_-_51802433 | 0.18 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr20_-_51802509 | 0.18 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr1_+_27935022 | 0.18 |
ENST00000411604.5
ENST00000373888.8 |
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr4_+_112860981 | 0.17 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112861053 | 0.17 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112860912 | 0.16 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr4_+_94974984 | 0.15 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr4_-_73620629 | 0.15 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr6_-_111606260 | 0.09 |
ENST00000340026.10
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr1_+_160190567 | 0.06 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr17_+_18183803 | 0.05 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr6_-_111605859 | 0.04 |
ENST00000651359.1
ENST00000650859.1 ENST00000359831.8 ENST00000368761.11 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr12_+_28257195 | 0.04 |
ENST00000381259.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_53336342 | 0.03 |
ENST00000537210.2
ENST00000536324.4 |
SP7
|
Sp7 transcription factor |
| chr17_-_42185452 | 0.02 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr21_-_31160904 | 0.01 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr20_+_31879798 | 0.01 |
ENST00000620043.4
|
TTLL9
|
tubulin tyrosine ligase like 9 |
| chr5_-_97142579 | 0.01 |
ENST00000274382.9
|
LIX1
|
limb and CNS expressed 1 |
| chr17_-_10026265 | 0.00 |
ENST00000437099.6
ENST00000396115.6 |
GAS7
|
growth arrest specific 7 |
| chr8_+_91249307 | 0.00 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EVX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |