Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for FOXM1_TBL1XR1
Z-value: 1.36
Transcription factors associated with FOXM1_TBL1XR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXM1
|
ENSG00000111206.13 | forkhead box M1 |
|
TBL1XR1
|
ENSG00000177565.18 | TBL1X receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBL1XR1 | hg38_v1_chr3_-_177197210_177197233 | -0.82 | 1.3e-02 | Click! |
| FOXM1 | hg38_v1_chr12_-_2876986_2877092, hg38_v1_chr12_-_2877113_2877182 | -0.81 | 1.5e-02 | Click! |
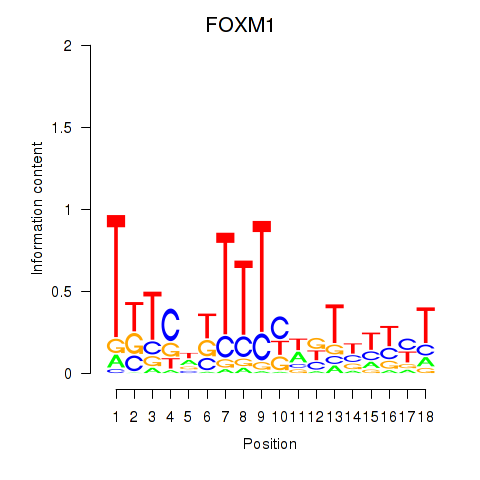
Activity profile of FOXM1_TBL1XR1 motif
Sorted Z-values of FOXM1_TBL1XR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_63887698 | 2.58 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr2_+_101991953 | 2.02 |
ENST00000332549.8
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr18_+_31447732 | 1.69 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr6_+_26500296 | 1.59 |
ENST00000684113.1
|
BTN1A1
|
butyrophilin subfamily 1 member A1 |
| chr19_-_50953093 | 1.53 |
ENST00000593428.5
|
KLK5
|
kallikrein related peptidase 5 |
| chr19_-_50953063 | 1.44 |
ENST00000391809.6
|
KLK5
|
kallikrein related peptidase 5 |
| chr2_+_102008515 | 1.25 |
ENST00000441002.1
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr19_-_50952942 | 1.24 |
ENST00000594846.1
ENST00000336334.8 |
KLK5
|
kallikrein related peptidase 5 |
| chr7_+_37848597 | 1.24 |
ENST00000199447.9
ENST00000455500.5 |
NME8
|
NME/NM23 family member 8 |
| chr11_-_102780620 | 1.13 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr12_-_8662073 | 1.12 |
ENST00000535411.5
ENST00000540087.5 |
MFAP5
|
microfibril associated protein 5 |
| chr11_+_5389377 | 1.10 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr12_-_8662703 | 1.09 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr10_-_104085847 | 1.05 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr18_+_23949847 | 1.02 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3 |
| chr5_+_140848360 | 1.01 |
ENST00000532602.2
|
PCDHA9
|
protocadherin alpha 9 |
| chr12_-_8662808 | 0.96 |
ENST00000359478.7
ENST00000396549.6 |
MFAP5
|
microfibril associated protein 5 |
| chr12_-_8662619 | 0.93 |
ENST00000544889.1
ENST00000543369.5 |
MFAP5
|
microfibril associated protein 5 |
| chr12_-_8650529 | 0.91 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr1_+_101238090 | 0.90 |
ENST00000475289.2
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr12_+_20695323 | 0.90 |
ENST00000266509.7
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr19_-_50968125 | 0.89 |
ENST00000594641.1
|
KLK6
|
kallikrein related peptidase 6 |
| chr2_-_112836702 | 0.87 |
ENST00000416750.1
ENST00000263341.7 ENST00000418817.5 |
IL1B
|
interleukin 1 beta |
| chr20_+_59604527 | 0.84 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr11_+_61222347 | 0.82 |
ENST00000378149.9
|
PGA4
|
pepsinogen A4 |
| chr8_-_41309434 | 0.81 |
ENST00000220772.8
|
SFRP1
|
secreted frizzled related protein 1 |
| chr19_-_50968775 | 0.81 |
ENST00000391808.5
|
KLK6
|
kallikrein related peptidase 6 |
| chr5_+_140841183 | 0.81 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr5_+_40841308 | 0.80 |
ENST00000381677.4
ENST00000254691.10 |
CARD6
|
caspase recruitment domain family member 6 |
| chr19_-_51020154 | 0.79 |
ENST00000391805.5
ENST00000599077.1 |
KLK10
|
kallikrein related peptidase 10 |
| chr18_+_63587336 | 0.77 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13 |
| chr15_+_43593054 | 0.77 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr12_+_50924005 | 0.76 |
ENST00000550502.1
|
METTL7A
|
methyltransferase like 7A |
| chr12_+_20695553 | 0.74 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr18_+_63587297 | 0.70 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr12_-_9999176 | 0.70 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr12_-_70637405 | 0.68 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr11_+_113908983 | 0.67 |
ENST00000537778.5
|
HTR3B
|
5-hydroxytryptamine receptor 3B |
| chr19_-_51020019 | 0.66 |
ENST00000309958.7
|
KLK10
|
kallikrein related peptidase 10 |
| chr1_+_161707244 | 0.66 |
ENST00000349527.8
ENST00000294796.8 ENST00000309691.10 ENST00000367953.7 ENST00000367950.2 |
FCRLA
|
Fc receptor like A |
| chr14_+_22112280 | 0.65 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr6_-_47042306 | 0.65 |
ENST00000371253.7
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr4_+_99816797 | 0.64 |
ENST00000512369.2
ENST00000296414.11 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr4_-_15938740 | 0.64 |
ENST00000382333.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr1_+_101237009 | 0.64 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr19_+_48270079 | 0.62 |
ENST00000595607.6
|
ZNF114
|
zinc finger protein 114 |
| chr14_+_22271921 | 0.62 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr20_-_45254556 | 0.62 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr1_+_160739286 | 0.61 |
ENST00000359331.8
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr6_+_28080565 | 0.60 |
ENST00000377325.2
ENST00000683778.1 |
ZNF165
|
zinc finger protein 165 |
| chr19_-_6720641 | 0.60 |
ENST00000245907.11
|
C3
|
complement C3 |
| chr3_-_172523460 | 0.60 |
ENST00000420541.6
|
TNFSF10
|
TNF superfamily member 10 |
| chr12_-_70609788 | 0.59 |
ENST00000547715.1
ENST00000538708.5 ENST00000550857.5 ENST00000261266.9 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr11_-_79441016 | 0.59 |
ENST00000278550.12
|
TENM4
|
teneurin transmembrane protein 4 |
| chr7_-_80922354 | 0.58 |
ENST00000419255.6
|
SEMA3C
|
semaphorin 3C |
| chr4_-_6070162 | 0.58 |
ENST00000636216.1
ENST00000637373.2 |
ENSG00000284684.1
JAKMIP1
|
novel protein janus kinase and microtubule interacting protein 1 |
| chr2_-_215138603 | 0.58 |
ENST00000272895.12
|
ABCA12
|
ATP binding cassette subfamily A member 12 |
| chr17_-_7263181 | 0.57 |
ENST00000575313.1
ENST00000397317.8 |
CLDN7
|
claudin 7 |
| chr1_-_28176665 | 0.57 |
ENST00000373857.8
|
PTAFR
|
platelet activating factor receptor |
| chr11_+_61203512 | 0.57 |
ENST00000325558.11
|
PGA3
|
pepsinogen A3 |
| chr8_+_32646838 | 0.57 |
ENST00000651333.1
ENST00000652592.1 |
NRG1
|
neuregulin 1 |
| chr19_+_49335396 | 0.57 |
ENST00000598095.5
ENST00000426897.6 ENST00000323906.9 ENST00000535669.6 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr19_-_50983815 | 0.57 |
ENST00000391807.5
ENST00000593904.1 ENST00000595820.6 |
KLK7
|
kallikrein related peptidase 7 |
| chr1_+_171090892 | 0.56 |
ENST00000367755.9
ENST00000479749.1 |
FMO3
|
flavin containing dimethylaniline monoxygenase 3 |
| chr1_-_12618198 | 0.56 |
ENST00000616661.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr6_+_30880780 | 0.55 |
ENST00000460944.6
ENST00000324771.12 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_+_5383812 | 0.55 |
ENST00000642046.1
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr11_-_5441514 | 0.55 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1 |
| chr1_+_160739265 | 0.55 |
ENST00000368042.7
|
SLAMF7
|
SLAM family member 7 |
| chr12_+_12891554 | 0.54 |
ENST00000014914.6
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A |
| chr8_-_56320098 | 0.54 |
ENST00000303749.8
ENST00000396721.6 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C member 5 |
| chr8_-_22693469 | 0.54 |
ENST00000317216.3
|
EGR3
|
early growth response 3 |
| chr10_+_97584314 | 0.54 |
ENST00000370647.8
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr19_+_35118456 | 0.53 |
ENST00000604621.5
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr5_+_140834230 | 0.53 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr12_+_8822610 | 0.53 |
ENST00000299698.12
|
A2ML1
|
alpha-2-macroglobulin like 1 |
| chr3_-_190122317 | 0.52 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr13_-_20230970 | 0.52 |
ENST00000644667.1
ENST00000646108.1 |
GJB6
|
gap junction protein beta 6 |
| chr1_+_24319342 | 0.52 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr1_+_161707222 | 0.52 |
ENST00000236938.12
|
FCRLA
|
Fc receptor like A |
| chr12_-_52493250 | 0.52 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr12_-_10674013 | 0.52 |
ENST00000535345.5
ENST00000542562.5 ENST00000075503.8 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr3_-_172523423 | 0.52 |
ENST00000241261.7
|
TNFSF10
|
TNF superfamily member 10 |
| chr2_+_102355881 | 0.51 |
ENST00000409599.5
|
IL18R1
|
interleukin 18 receptor 1 |
| chr8_+_133113483 | 0.51 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr18_-_55635948 | 0.51 |
ENST00000565124.4
ENST00000398339.5 |
TCF4
|
transcription factor 4 |
| chr1_+_89364051 | 0.51 |
ENST00000370456.5
|
GBP6
|
guanylate binding protein family member 6 |
| chr3_+_97439603 | 0.51 |
ENST00000514100.5
|
EPHA6
|
EPH receptor A6 |
| chr1_+_86424154 | 0.50 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2 |
| chr2_+_102355750 | 0.50 |
ENST00000233957.7
|
IL18R1
|
interleukin 18 receptor 1 |
| chr4_+_40192949 | 0.50 |
ENST00000507851.5
ENST00000615577.4 ENST00000613272.4 |
RHOH
|
ras homolog family member H |
| chr6_+_30888672 | 0.49 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_68734773 | 0.49 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr16_+_75222609 | 0.49 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr2_-_31414694 | 0.49 |
ENST00000379416.4
|
XDH
|
xanthine dehydrogenase |
| chr7_-_36367141 | 0.48 |
ENST00000453212.5
ENST00000415803.2 ENST00000431396.1 ENST00000440378.6 ENST00000317020.10 ENST00000436884.5 |
KIAA0895
|
KIAA0895 |
| chr6_+_111259474 | 0.48 |
ENST00000672554.1
ENST00000673024.1 |
MFSD4B
|
major facilitator superfamily domain containing 4B |
| chr5_+_36606355 | 0.48 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr7_+_16753731 | 0.48 |
ENST00000262067.5
|
TSPAN13
|
tetraspanin 13 |
| chr9_+_5450503 | 0.47 |
ENST00000381573.8
ENST00000381577.4 |
CD274
|
CD274 molecule |
| chr1_+_158180863 | 0.47 |
ENST00000673623.2
ENST00000674085.2 ENST00000673701.2 |
CD1D
|
CD1d molecule |
| chr1_+_161707205 | 0.47 |
ENST00000367957.7
|
FCRLA
|
Fc receptor like A |
| chr12_-_8662881 | 0.47 |
ENST00000433590.6
|
MFAP5
|
microfibril associated protein 5 |
| chr9_+_27524285 | 0.47 |
ENST00000276943.3
|
IFNK
|
interferon kappa |
| chrX_+_136532205 | 0.47 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1 |
| chr17_-_7263959 | 0.46 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr2_+_102311502 | 0.46 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr8_+_132866948 | 0.46 |
ENST00000220616.9
|
TG
|
thyroglobulin |
| chr19_-_51019699 | 0.46 |
ENST00000358789.8
|
KLK10
|
kallikrein related peptidase 10 |
| chr5_+_36608146 | 0.46 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr12_+_107318395 | 0.46 |
ENST00000420571.6
ENST00000280758.10 |
BTBD11
|
BTB domain containing 11 |
| chr11_-_120123026 | 0.46 |
ENST00000533302.5
|
TRIM29
|
tripartite motif containing 29 |
| chr18_-_31102411 | 0.46 |
ENST00000251081.8
ENST00000280904.11 ENST00000682357.1 ENST00000648081.1 |
DSC2
|
desmocollin 2 |
| chr12_-_6631632 | 0.45 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr6_+_26383090 | 0.45 |
ENST00000469230.5
ENST00000490025.5 ENST00000352867.6 ENST00000356709.9 ENST00000493275.5 ENST00000472507.5 ENST00000482536.5 ENST00000432533.6 ENST00000482842.1 |
BTN2A2
|
butyrophilin subfamily 2 member A2 |
| chr14_-_94390650 | 0.45 |
ENST00000449399.7
ENST00000404814.8 |
SERPINA1
|
serpin family A member 1 |
| chr14_+_23258485 | 0.45 |
ENST00000399905.5
ENST00000470456.1 |
RNF212B
|
ring finger protein 212B |
| chr5_-_16508812 | 0.45 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr4_-_56821679 | 0.44 |
ENST00000504762.1
ENST00000506738.6 ENST00000248701.8 ENST00000616980.1 ENST00000618802.3 ENST00000631082.1 |
SPINK2
|
serine peptidase inhibitor Kazal type 2 |
| chr5_+_140786136 | 0.44 |
ENST00000378133.4
ENST00000504120.4 |
PCDHA1
|
protocadherin alpha 1 |
| chr1_-_161021096 | 0.44 |
ENST00000537746.1
ENST00000368026.11 |
F11R
|
F11 receptor |
| chr1_+_34755039 | 0.43 |
ENST00000338513.1
|
GJB5
|
gap junction protein beta 5 |
| chr6_-_132798587 | 0.43 |
ENST00000275227.9
|
SLC18B1
|
solute carrier family 18 member B1 |
| chr4_+_40196907 | 0.43 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr3_-_190322434 | 0.43 |
ENST00000295522.4
|
CLDN1
|
claudin 1 |
| chr17_+_7630094 | 0.43 |
ENST00000441599.6
ENST00000380450.9 ENST00000416273.7 ENST00000575903.5 ENST00000571153.5 ENST00000575618.5 ENST00000576152.1 ENST00000576830.5 |
SHBG
|
sex hormone binding globulin |
| chr14_+_21042352 | 0.43 |
ENST00000298690.5
|
RNASE7
|
ribonuclease A family member 7 |
| chr11_-_60952067 | 0.42 |
ENST00000681275.1
|
SLC15A3
|
solute carrier family 15 member 3 |
| chr19_-_14979676 | 0.42 |
ENST00000598504.5
ENST00000597262.1 |
SLC1A6
|
solute carrier family 1 member 6 |
| chr4_-_88231322 | 0.42 |
ENST00000515655.5
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr12_-_8066331 | 0.42 |
ENST00000546241.1
ENST00000307637.5 |
C3AR1
|
complement C3a receptor 1 |
| chr8_+_94641074 | 0.42 |
ENST00000423620.6
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr5_+_140786291 | 0.42 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1 |
| chr12_+_41437680 | 0.42 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr4_-_99352754 | 0.42 |
ENST00000639454.1
|
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr4_+_87975829 | 0.41 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr18_+_31376777 | 0.41 |
ENST00000308128.9
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr18_+_23873000 | 0.41 |
ENST00000269217.11
ENST00000587184.5 |
LAMA3
|
laminin subunit alpha 3 |
| chr10_+_116427839 | 0.41 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr6_+_130421086 | 0.41 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A |
| chr19_-_51027954 | 0.41 |
ENST00000391804.7
|
KLK11
|
kallikrein related peptidase 11 |
| chr14_+_73097027 | 0.41 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr9_+_21409116 | 0.41 |
ENST00000380205.2
|
IFNA8
|
interferon alpha 8 |
| chr7_-_143408848 | 0.41 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1 |
| chr19_-_50969567 | 0.41 |
ENST00000310157.7
|
KLK6
|
kallikrein related peptidase 6 |
| chr19_+_9250930 | 0.41 |
ENST00000456448.3
|
OR7E24
|
olfactory receptor family 7 subfamily E member 24 |
| chr17_-_40937641 | 0.41 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr18_+_63907948 | 0.40 |
ENST00000238508.8
|
SERPINB10
|
serpin family B member 10 |
| chr6_-_116060859 | 0.40 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr3_-_55489938 | 0.40 |
ENST00000474267.5
|
WNT5A
|
Wnt family member 5A |
| chr1_+_86547070 | 0.40 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr5_+_140875299 | 0.40 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chr1_+_196774813 | 0.40 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr12_+_75481204 | 0.40 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr17_-_58415628 | 0.40 |
ENST00000583753.5
|
RNF43
|
ring finger protein 43 |
| chr12_+_75480800 | 0.39 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr14_-_99271485 | 0.39 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr7_+_117020191 | 0.39 |
ENST00000434836.5
ENST00000393443.5 ENST00000465133.5 ENST00000477742.5 ENST00000393444.7 ENST00000393447.8 |
ST7
|
suppression of tumorigenicity 7 |
| chr5_-_16508788 | 0.39 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr1_+_34782259 | 0.39 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chr12_-_66678934 | 0.39 |
ENST00000545666.5
ENST00000398016.7 ENST00000359742.9 ENST00000538211.5 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr3_+_189789643 | 0.39 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chr11_+_85855377 | 0.39 |
ENST00000342404.8
|
CCDC83
|
coiled-coil domain containing 83 |
| chr18_-_55322215 | 0.39 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr11_-_120128831 | 0.39 |
ENST00000529044.5
|
TRIM29
|
tripartite motif containing 29 |
| chr11_+_34621065 | 0.39 |
ENST00000257831.8
|
EHF
|
ETS homologous factor |
| chr9_-_41908681 | 0.39 |
ENST00000476961.5
|
CNTNAP3B
|
contactin associated protein family member 3B |
| chr4_-_11428868 | 0.38 |
ENST00000002596.6
|
HS3ST1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr6_+_26383176 | 0.38 |
ENST00000416795.6
ENST00000494184.1 |
BTN2A2
|
butyrophilin subfamily 2 member A2 |
| chr12_-_27970047 | 0.38 |
ENST00000395868.7
|
PTHLH
|
parathyroid hormone like hormone |
| chr10_+_89332484 | 0.38 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr7_-_41700583 | 0.38 |
ENST00000442711.1
|
INHBA
|
inhibin subunit beta A |
| chrX_-_133097095 | 0.38 |
ENST00000511190.5
|
USP26
|
ubiquitin specific peptidase 26 |
| chr1_-_153615858 | 0.38 |
ENST00000476873.5
|
S100A14
|
S100 calcium binding protein A14 |
| chr9_-_127778659 | 0.38 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C |
| chr19_+_41797147 | 0.38 |
ENST00000596544.1
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr1_-_169711603 | 0.38 |
ENST00000236147.6
ENST00000650983.1 |
SELL
|
selectin L |
| chr13_-_46182136 | 0.38 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr9_-_104928139 | 0.38 |
ENST00000423487.6
ENST00000374733.1 ENST00000374736.8 ENST00000678995.1 |
ABCA1
|
ATP binding cassette subfamily A member 1 |
| chr9_-_114806031 | 0.38 |
ENST00000374045.5
|
TNFSF15
|
TNF superfamily member 15 |
| chr19_-_51028015 | 0.38 |
ENST00000319720.11
|
KLK11
|
kallikrein related peptidase 11 |
| chr12_+_40692413 | 0.38 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr12_-_11062294 | 0.38 |
ENST00000533467.1
|
TAS2R46
|
taste 2 receptor member 46 |
| chr8_+_94641145 | 0.37 |
ENST00000433389.8
ENST00000358397.9 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr4_+_47485268 | 0.37 |
ENST00000273859.8
ENST00000504445.1 |
ATP10D
|
ATPase phospholipid transporting 10D (putative) |
| chr4_+_73869385 | 0.37 |
ENST00000395761.4
|
CXCL1
|
C-X-C motif chemokine ligand 1 |
| chr6_-_39229465 | 0.37 |
ENST00000359534.4
|
KCNK5
|
potassium two pore domain channel subfamily K member 5 |
| chr19_-_42877988 | 0.37 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr17_+_7455571 | 0.37 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr11_+_18132565 | 0.37 |
ENST00000621697.2
|
MRGPRX3
|
MAS related GPR family member X3 |
| chr6_+_112054075 | 0.37 |
ENST00000230529.9
ENST00000604763.5 ENST00000368666.7 ENST00000674325.1 ENST00000483439.1 ENST00000409166.5 |
CCN6
|
cellular communication network factor 6 |
| chr19_-_54173151 | 0.37 |
ENST00000619895.5
|
TMC4
|
transmembrane channel like 4 |
| chr19_-_50984028 | 0.37 |
ENST00000597707.5
|
KLK7
|
kallikrein related peptidase 7 |
| chr10_+_89392546 | 0.36 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr12_-_54981838 | 0.36 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr16_+_5033923 | 0.36 |
ENST00000683433.1
ENST00000682020.1 ENST00000682985.1 ENST00000682327.1 ENST00000588623.5 |
ALG1
|
ALG1 chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr10_+_89301932 | 0.36 |
ENST00000371826.4
ENST00000679755.1 |
IFIT2
|
interferon induced protein with tetratricopeptide repeats 2 |
| chr6_+_121435595 | 0.36 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr3_-_116444983 | 0.36 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr6_-_2841853 | 0.36 |
ENST00000380739.6
|
SERPINB1
|
serpin family B member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXM1_TBL1XR1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.9 | 5.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.5 | 1.5 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 1.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.3 | 1.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 | 1.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 1.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 0.7 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 1.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 0.6 | GO:0002892 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.2 | 2.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.2 | 0.8 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 0.8 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.2 | 0.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.7 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 | 0.5 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 0.5 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.2 | 0.6 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.2 | 0.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.6 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 0.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 0.9 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 0.9 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.1 | 0.7 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.4 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 0.1 | 0.7 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.6 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.1 | GO:0002436 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.1 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.4 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.4 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 0.8 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 1.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.4 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 1.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.4 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.1 | 1.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 | 0.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.4 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.1 | 0.6 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.1 | 0.6 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 1.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.2 | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.1 | 3.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.1 | 0.7 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 1.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 3.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 5.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.4 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.3 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.1 | 0.7 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.1 | 0.3 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.3 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.1 | 0.3 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.4 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.3 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 0.3 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.8 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.5 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.1 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.1 | 0.7 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.3 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.5 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.4 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.2 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 2.1 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.3 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.4 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.1 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.3 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.3 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 1.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.8 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.5 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.1 | 0.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.8 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) negative regulation of pinocytosis(GO:0048550) |
| 0.1 | 0.4 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.9 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.5 | GO:0019883 | antigen processing and presentation of endogenous antigen(GO:0019883) |
| 0.1 | 0.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.9 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.9 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.5 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.4 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.1 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.1 | 0.2 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 0.2 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 2.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.1 | 8.2 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.1 | GO:0042214 | carotene metabolic process(GO:0016119) terpene metabolic process(GO:0042214) |
| 0.1 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 0.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.5 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.1 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 0.2 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.9 | GO:0002638 | negative regulation of immunoglobulin production(GO:0002638) |
| 0.1 | 0.1 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 | 0.2 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.4 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.4 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 0.2 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.1 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.1 | 0.1 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.5 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.8 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.2 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.1 | 0.2 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.1 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 1.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.0 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.4 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.1 | 0.1 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.1 | 0.2 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.2 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 0.2 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.2 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.0 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.4 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.5 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 1.8 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0031393 | negative regulation of prostaglandin biosynthetic process(GO:0031393) |
| 0.0 | 0.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.0 | 0.1 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0051133 | regulation of NK T cell activation(GO:0051133) positive regulation of NK T cell activation(GO:0051135) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 1.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.5 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.3 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.1 | GO:0060266 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.5 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.3 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.5 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) |
| 0.0 | 0.4 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 1.4 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.9 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.3 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 6.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0060356 | tryptophan transport(GO:0015827) leucine import(GO:0060356) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 2.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.8 | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.6 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.2 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.4 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:1902164 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.5 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:2000020 | regulation of male gonad development(GO:2000018) positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.1 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 1.3 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.1 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.2 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.3 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.3 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0036047 | protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) peptidyl-lysine deglutarylation(GO:0061699) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0010513 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.0 | GO:0030656 | regulation of vitamin metabolic process(GO:0030656) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.2 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.4 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.0 | 0.0 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.0 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.7 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.7 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.0 | GO:2000848 | positive regulation of corticosteroid hormone secretion(GO:2000848) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.0 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.3 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0033168 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 0.0 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.0 | 0.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.0 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.4 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) |
| 0.0 | 0.2 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0070650 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.0 | GO:0097695 | establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0033079 | immature T cell proliferation(GO:0033079) regulation of immature T cell proliferation(GO:0033083) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.1 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.0 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0070091 | glucagon secretion(GO:0070091) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.0 | GO:0002583 | regulation of antigen processing and presentation of peptide antigen(GO:0002583) |
| 0.0 | 1.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.4 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.0 | GO:0001812 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0009111 | vitamin catabolic process(GO:0009111) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.0 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.0 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0060456 | positive regulation of digestive system process(GO:0060456) |
| 0.0 | 0.0 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) |
| 0.0 | 0.0 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0046629 | gamma-delta T cell activation(GO:0046629) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.0 | GO:0002396 | MHC protein complex assembly(GO:0002396) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 2.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.3 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.0 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) cytolysis in other organism involved in symbiotic interaction(GO:0051801) |
| 0.0 | 0.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.0 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.0 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0032849 | maternal aggressive behavior(GO:0002125) positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0045556 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.0 | 1.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0061197 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.0 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.0 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.0 | 0.0 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.0 | 0.1 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.0 | 0.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0018364 | box C/D snoRNA 3'-end processing(GO:0000494) peptidyl-glutamine methylation(GO:0018364) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.4 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.0 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.0 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.0 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.0 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0071476 | hypotonic response(GO:0006971) cellular hypotonic response(GO:0071476) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.8 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 1.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 5.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 0.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 0.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 1.8 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 0.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.4 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.5 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 5.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.3 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.7 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 1.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 1.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.2 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 0.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.2 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.1 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.2 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.0 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 3.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 2.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0033167 | ARC complex(GO:0033167) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 1.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 0.0 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.2 | GO:0060198 | clathrin-sculpted vesicle(GO:0060198) clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 5.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 78.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.5 | 1.5 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.7 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.2 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.5 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.2 | 3.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.6 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.2 | 0.6 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 2.0 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 1.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.3 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.1 | 0.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 1.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.9 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 1.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.1 | GO:0035673 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.7 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.2 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.1 | 0.6 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.3 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.1 | 0.4 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.1 | 0.8 | GO:0046978 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.5 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 2.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.5 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.1 | 1.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.9 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.5 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.2 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.1 | 0.5 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.7 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 1.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.4 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 0.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 1.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.3 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 8.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 2.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 1.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 1.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.2 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.1 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.1 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 1.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.3 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.4 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 13.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.5 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.0 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.8 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 4.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 1.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 1.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0016154 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.0 | 0.1 | GO:0036054 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0010851 | cyclase regulator activity(GO:0010851) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.0 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.0 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.0 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 4.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 10.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 4.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.5 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 2.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 4.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.1 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |