Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
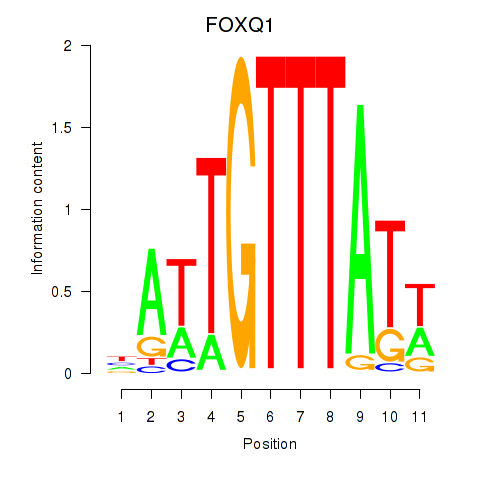
Results for FOXQ1
Z-value: 1.07
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.7 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg38_v1_chr6_+_1312090_1312105 | -0.84 | 9.1e-03 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91180365 | 2.92 |
ENST00000547937.5
|
DCN
|
decorin |
| chr2_+_151357583 | 2.01 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr3_+_157436842 | 1.93 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr2_-_187554351 | 1.55 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr1_-_72100930 | 1.52 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr5_-_20575850 | 1.46 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr5_+_141150012 | 1.39 |
ENST00000231136.4
ENST00000622991.1 |
PCDHB6
|
protocadherin beta 6 |
| chr4_-_69760596 | 1.25 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr1_-_56579555 | 1.15 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr2_-_189179754 | 1.12 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr4_-_69760610 | 1.11 |
ENST00000310613.8
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr20_+_13008919 | 0.98 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr10_-_77637444 | 0.95 |
ENST00000639205.1
ENST00000639498.1 ENST00000372408.7 ENST00000372403.9 ENST00000640626.1 ENST00000404857.6 ENST00000638252.1 ENST00000640029.1 ENST00000640934.1 ENST00000639823.1 ENST00000372437.6 ENST00000639344.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr3_+_69936583 | 0.94 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr1_-_145910031 | 0.88 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10 |
| chr14_+_75632610 | 0.87 |
ENST00000555027.1
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr8_+_78516329 | 0.87 |
ENST00000396418.7
ENST00000352966.9 |
PKIA
|
cAMP-dependent protein kinase inhibitor alpha |
| chr9_-_20382461 | 0.83 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr1_-_145910066 | 0.82 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10 |
| chr6_-_87095059 | 0.78 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr12_+_1629197 | 0.77 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B |
| chr5_-_111756245 | 0.77 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr7_+_93921720 | 0.74 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr8_+_69492793 | 0.73 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr12_-_15221394 | 0.68 |
ENST00000537647.5
ENST00000256953.6 ENST00000546331.5 |
RERG
|
RAS like estrogen regulated growth inhibitor |
| chrX_+_9912434 | 0.65 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr12_-_10453330 | 0.64 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr20_+_36214373 | 0.63 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr5_+_141192330 | 0.63 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr18_-_27990256 | 0.60 |
ENST00000675173.1
|
CDH2
|
cadherin 2 |
| chr5_-_41510623 | 0.60 |
ENST00000328457.5
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3 |
| chr1_+_162632454 | 0.57 |
ENST00000367921.8
ENST00000367922.7 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr3_+_155083889 | 0.57 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr9_-_121050264 | 0.56 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr1_-_27672178 | 0.55 |
ENST00000339145.8
ENST00000361157.11 ENST00000362020.4 ENST00000679644.1 |
IFI6
|
interferon alpha inducible protein 6 |
| chr4_-_99352754 | 0.55 |
ENST00000639454.1
|
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr1_-_23868279 | 0.54 |
ENST00000374479.4
|
FUCA1
|
alpha-L-fucosidase 1 |
| chr15_+_41256907 | 0.54 |
ENST00000560965.1
|
CHP1
|
calcineurin like EF-hand protein 1 |
| chr2_-_187554473 | 0.54 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr19_+_49527988 | 0.52 |
ENST00000270645.8
|
RCN3
|
reticulocalbin 3 |
| chr10_-_48604952 | 0.51 |
ENST00000417912.6
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr18_+_7754959 | 0.51 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr2_-_144517663 | 0.51 |
ENST00000427902.5
ENST00000462355.2 ENST00000470879.5 ENST00000409487.7 ENST00000435831.5 ENST00000630572.2 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_141364153 | 0.51 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr4_-_118352967 | 0.49 |
ENST00000296498.3
|
PRSS12
|
serine protease 12 |
| chr19_+_926001 | 0.48 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr1_-_53940100 | 0.48 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr5_-_41510554 | 0.48 |
ENST00000377801.8
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3 |
| chr3_+_155083523 | 0.48 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr17_-_445939 | 0.48 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr7_+_134843884 | 0.47 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr12_-_71157872 | 0.46 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr4_-_158173042 | 0.46 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr14_+_88005128 | 0.46 |
ENST00000267549.5
|
GPR65
|
G protein-coupled receptor 65 |
| chr10_-_48605032 | 0.46 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chrX_+_10158448 | 0.46 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr4_-_158173004 | 0.45 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr3_+_152300135 | 0.44 |
ENST00000465907.6
ENST00000492948.5 ENST00000485509.5 ENST00000464596.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr5_-_88785493 | 0.42 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr14_-_73027077 | 0.41 |
ENST00000553891.5
ENST00000556143.6 |
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr1_+_61077219 | 0.41 |
ENST00000407417.7
|
NFIA
|
nuclear factor I A |
| chr15_-_63157464 | 0.40 |
ENST00000330964.10
ENST00000635699.1 ENST00000439025.1 |
RPS27L
|
ribosomal protein S27 like |
| chr2_-_202871253 | 0.39 |
ENST00000435143.5
|
ICA1L
|
islet cell autoantigen 1 like |
| chr4_-_185775271 | 0.39 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_120477354 | 0.39 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chrX_-_100410264 | 0.39 |
ENST00000373034.8
|
PCDH19
|
protocadherin 19 |
| chr12_+_18262730 | 0.37 |
ENST00000675017.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr5_+_141364231 | 0.37 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr11_+_5689780 | 0.37 |
ENST00000379965.8
ENST00000454828.5 |
TRIM22
|
tripartite motif containing 22 |
| chr14_+_50533026 | 0.37 |
ENST00000441560.6
|
ATL1
|
atlastin GTPase 1 |
| chr19_+_49930219 | 0.36 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr4_-_139280179 | 0.36 |
ENST00000398955.2
|
MGARP
|
mitochondria localized glutamic acid rich protein |
| chr7_-_139716980 | 0.36 |
ENST00000342645.7
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr2_+_200440649 | 0.36 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr14_-_73027117 | 0.35 |
ENST00000318876.9
|
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr19_-_58353482 | 0.34 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr1_+_103655760 | 0.34 |
ENST00000370083.9
|
AMY1A
|
amylase alpha 1A |
| chr12_-_122395422 | 0.33 |
ENST00000540304.6
|
CLIP1
|
CAP-Gly domain containing linker protein 1 |
| chr6_+_135851681 | 0.33 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr1_+_171090892 | 0.33 |
ENST00000367755.9
ENST00000479749.1 |
FMO3
|
flavin containing dimethylaniline monoxygenase 3 |
| chr11_+_20022550 | 0.32 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr7_+_143288215 | 0.32 |
ENST00000619992.4
ENST00000310447.10 |
CASP2
|
caspase 2 |
| chr5_+_141338753 | 0.31 |
ENST00000528330.2
ENST00000394576.3 |
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr2_+_108621260 | 0.30 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr2_-_105438503 | 0.30 |
ENST00000393352.7
ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr7_-_138627444 | 0.30 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr5_-_88823763 | 0.30 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr15_-_55365231 | 0.30 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr6_+_132570322 | 0.29 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr14_+_50560137 | 0.29 |
ENST00000358385.12
|
ATL1
|
atlastin GTPase 1 |
| chr4_+_38867677 | 0.29 |
ENST00000510213.5
ENST00000515037.5 |
FAM114A1
|
family with sequence similarity 114 member A1 |
| chr16_-_31065011 | 0.29 |
ENST00000539836.3
ENST00000535577.5 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr3_-_114624193 | 0.28 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_174875505 | 0.28 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr19_-_10335773 | 0.28 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr1_+_103617427 | 0.28 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr11_-_124800630 | 0.28 |
ENST00000239614.8
ENST00000674284.1 |
MSANTD2
|
Myb/SANT DNA binding domain containing 2 |
| chr5_+_141245384 | 0.27 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chr11_+_73308237 | 0.27 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr3_-_114759115 | 0.26 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_103749898 | 0.26 |
ENST00000622339.5
|
AMY1C
|
amylase alpha 1C |
| chr18_+_68798065 | 0.26 |
ENST00000360242.9
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr14_-_73950393 | 0.25 |
ENST00000651776.1
|
FAM161B
|
FAM161 centrosomal protein B |
| chr10_+_79703227 | 0.25 |
ENST00000429828.5
ENST00000372321.5 |
NUTM2B
|
NUT family member 2B |
| chr10_+_15043937 | 0.25 |
ENST00000378228.8
ENST00000378217.3 |
OLAH
|
oleoyl-ACP hydrolase |
| chr5_+_72848161 | 0.25 |
ENST00000506351.6
|
TNPO1
|
transportin 1 |
| chr11_+_65945445 | 0.23 |
ENST00000532620.5
|
TSGA10IP
|
testis specific 10 interacting protein |
| chr20_+_58389197 | 0.23 |
ENST00000475243.6
ENST00000395802.7 |
VAPB
|
VAMP associated protein B and C |
| chr20_-_45667177 | 0.23 |
ENST00000618797.4
|
WFDC11
|
WAP four-disulfide core domain 11 |
| chr14_-_89412025 | 0.23 |
ENST00000553840.5
ENST00000556916.5 |
FOXN3
|
forkhead box N3 |
| chr14_-_106811131 | 0.23 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr5_+_72848115 | 0.23 |
ENST00000679378.1
|
TNPO1
|
transportin 1 |
| chr1_+_60865259 | 0.22 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr15_+_91100194 | 0.22 |
ENST00000394232.6
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr3_-_179259208 | 0.22 |
ENST00000485523.5
|
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr12_-_71157992 | 0.22 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8 |
| chr10_-_91909476 | 0.22 |
ENST00000311575.6
|
FGFBP3
|
fibroblast growth factor binding protein 3 |
| chr16_+_22490337 | 0.21 |
ENST00000415833.6
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chrX_-_72305892 | 0.21 |
ENST00000450875.5
ENST00000417400.1 ENST00000431381.5 ENST00000445983.5 ENST00000651998.1 |
CITED1
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1 |
| chr5_+_141208697 | 0.21 |
ENST00000624949.1
ENST00000622978.1 ENST00000239450.4 |
PCDHB12
|
protocadherin beta 12 |
| chr20_+_49812697 | 0.21 |
ENST00000417961.5
|
SLC9A8
|
solute carrier family 9 member A8 |
| chr7_+_28412511 | 0.20 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr1_+_74235377 | 0.20 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr20_+_31514410 | 0.20 |
ENST00000335574.10
ENST00000340852.9 ENST00000398174.9 ENST00000466766.2 ENST00000498035.5 ENST00000344042.5 |
HM13
|
histocompatibility minor 13 |
| chr11_-_55936400 | 0.20 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr2_-_202870862 | 0.20 |
ENST00000454326.5
ENST00000432273.5 ENST00000450143.5 ENST00000411681.5 |
ICA1L
|
islet cell autoantigen 1 like |
| chr11_+_72080313 | 0.20 |
ENST00000307198.11
ENST00000538413.6 ENST00000642648.1 ENST00000289488.7 |
ENSG00000284922.2
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chr17_-_48613468 | 0.20 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr3_+_69936629 | 0.20 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr1_+_236523417 | 0.19 |
ENST00000341872.10
ENST00000416919.6 ENST00000450372.6 |
LGALS8
|
galectin 8 |
| chr6_-_130956371 | 0.19 |
ENST00000639623.1
ENST00000525193.5 ENST00000527659.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr16_+_4734519 | 0.19 |
ENST00000299320.10
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr15_-_56465130 | 0.19 |
ENST00000260453.4
|
MNS1
|
meiosis specific nuclear structural 1 |
| chr5_-_131796921 | 0.19 |
ENST00000307968.11
ENST00000307954.12 |
FNIP1
|
folliculin interacting protein 1 |
| chr14_-_91732059 | 0.18 |
ENST00000553329.5
ENST00000256343.8 |
CATSPERB
|
cation channel sperm associated auxiliary subunit beta |
| chr11_-_47426419 | 0.18 |
ENST00000298852.8
ENST00000530912.5 ENST00000619920.4 |
PSMC3
|
proteasome 26S subunit, ATPase 3 |
| chr19_-_7294406 | 0.18 |
ENST00000302850.10
|
INSR
|
insulin receptor |
| chrX_+_109535775 | 0.18 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr1_+_84164962 | 0.18 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr1_+_27773189 | 0.18 |
ENST00000373943.9
ENST00000440806.2 |
STX12
|
syntaxin 12 |
| chr15_+_31366138 | 0.18 |
ENST00000558844.1
|
KLF13
|
Kruppel like factor 13 |
| chr17_-_41065879 | 0.18 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chrX_+_88747225 | 0.18 |
ENST00000276127.9
ENST00000373111.5 |
CPXCR1
|
CPX chromosome region candidate 1 |
| chr7_+_48171451 | 0.18 |
ENST00000435803.6
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr16_+_14708944 | 0.18 |
ENST00000526520.5
ENST00000531598.6 |
NPIPA3
|
nuclear pore complex interacting protein family member A3 |
| chr5_+_136132772 | 0.18 |
ENST00000545279.6
ENST00000507118.5 ENST00000511116.5 ENST00000545620.5 ENST00000509297.6 |
SMAD5
|
SMAD family member 5 |
| chr5_-_160419059 | 0.18 |
ENST00000297151.9
ENST00000519349.5 ENST00000520664.1 |
SLU7
|
SLU7 homolog, splicing factor |
| chr12_-_49189053 | 0.17 |
ENST00000550767.6
ENST00000546918.1 ENST00000679733.1 ENST00000552924.2 ENST00000301071.12 |
TUBA1A
|
tubulin alpha 1a |
| chr6_-_29005313 | 0.17 |
ENST00000377179.4
|
ZNF311
|
zinc finger protein 311 |
| chr21_+_32298849 | 0.17 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr11_+_30323077 | 0.17 |
ENST00000282032.4
|
ARL14EP
|
ADP ribosylation factor like GTPase 14 effector protein |
| chr2_+_85354720 | 0.17 |
ENST00000409331.6
ENST00000409344.7 ENST00000393852.8 ENST00000409013.8 ENST00000315658.11 ENST00000462891.7 |
ELMOD3
|
ELMO domain containing 3 |
| chr3_+_107377433 | 0.17 |
ENST00000261058.3
|
CCDC54
|
coiled-coil domain containing 54 |
| chr13_-_38990824 | 0.17 |
ENST00000379631.9
|
STOML3
|
stomatin like 3 |
| chr1_+_220748297 | 0.17 |
ENST00000366913.8
|
MTARC2
|
mitochondrial amidoxime reducing component 2 |
| chr10_+_35195843 | 0.17 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr1_+_196977550 | 0.16 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr12_-_91111460 | 0.16 |
ENST00000266718.5
|
LUM
|
lumican |
| chr10_-_103153609 | 0.16 |
ENST00000675985.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr13_-_38990856 | 0.16 |
ENST00000423210.1
|
STOML3
|
stomatin like 3 |
| chrX_-_119943732 | 0.16 |
ENST00000371410.5
|
NKAP
|
NFKB activating protein |
| chr10_-_30999469 | 0.16 |
ENST00000538351.6
|
ZNF438
|
zinc finger protein 438 |
| chr5_+_119476530 | 0.16 |
ENST00000645099.1
ENST00000513628.5 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr10_+_80131660 | 0.16 |
ENST00000372270.6
|
PLAC9
|
placenta associated 9 |
| chr12_-_26833143 | 0.16 |
ENST00000381340.8
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor type 2 |
| chr12_+_109116581 | 0.16 |
ENST00000338432.11
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr1_+_84164370 | 0.15 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr1_-_94925759 | 0.15 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr20_-_3204633 | 0.15 |
ENST00000354488.8
ENST00000380201.2 |
DDRGK1
|
DDRGK domain containing 1 |
| chr19_+_57633161 | 0.15 |
ENST00000541801.5
ENST00000347302.7 ENST00000240731.5 ENST00000254182.11 ENST00000391703.3 |
ZNF211
|
zinc finger protein 211 |
| chr3_+_156674579 | 0.15 |
ENST00000295924.12
|
TIPARP
|
TCDD inducible poly(ADP-ribose) polymerase |
| chr16_+_4734457 | 0.15 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr1_+_113929600 | 0.15 |
ENST00000369558.5
ENST00000369561.8 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr10_+_70404129 | 0.15 |
ENST00000373218.5
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr1_-_70354673 | 0.15 |
ENST00000370944.9
ENST00000262346.6 |
ANKRD13C
|
ankyrin repeat domain 13C |
| chr3_+_63967738 | 0.15 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr9_+_69820827 | 0.15 |
ENST00000527647.5
ENST00000480564.1 |
C9orf135
|
chromosome 9 open reading frame 135 |
| chr22_-_50245016 | 0.15 |
ENST00000248846.10
|
TUBGCP6
|
tubulin gamma complex associated protein 6 |
| chr4_-_21948733 | 0.15 |
ENST00000447367.6
ENST00000382152.7 |
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr8_+_7836433 | 0.14 |
ENST00000314265.3
|
DEFB104A
|
defensin beta 104A |
| chr10_+_21524670 | 0.14 |
ENST00000631589.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr12_+_121132869 | 0.14 |
ENST00000328963.10
|
P2RX7
|
purinergic receptor P2X 7 |
| chr11_-_615570 | 0.14 |
ENST00000649187.1
ENST00000647801.1 ENST00000397566.5 ENST00000397570.5 |
IRF7
|
interferon regulatory factor 7 |
| chr11_+_65027402 | 0.14 |
ENST00000377244.8
ENST00000534637.5 ENST00000524831.5 |
SNX15
|
sorting nexin 15 |
| chr19_+_36687579 | 0.14 |
ENST00000682579.1
ENST00000536254.6 |
ZNF567
|
zinc finger protein 567 |
| chr3_+_184812138 | 0.14 |
ENST00000287546.8
|
VPS8
|
VPS8 subunit of CORVET complex |
| chr2_+_73892967 | 0.14 |
ENST00000409731.7
ENST00000409918.5 ENST00000442912.5 ENST00000345517.8 ENST00000409624.1 |
ACTG2
|
actin gamma 2, smooth muscle |
| chr3_+_184812159 | 0.14 |
ENST00000625842.3
ENST00000436792.6 ENST00000446204.6 ENST00000422105.5 |
VPS8
|
VPS8 subunit of CORVET complex |
| chr14_-_60724300 | 0.14 |
ENST00000556952.3
ENST00000216513.5 |
SIX4
|
SIX homeobox 4 |
| chr2_-_174764407 | 0.14 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr14_-_106627685 | 0.14 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chrX_-_109482075 | 0.14 |
ENST00000218006.3
|
GUCY2F
|
guanylate cyclase 2F, retinal |
| chr10_+_35195124 | 0.14 |
ENST00000487763.5
ENST00000473940.5 ENST00000488328.5 ENST00000356917.9 |
CREM
|
cAMP responsive element modulator |
| chr13_+_75804270 | 0.14 |
ENST00000447038.5
|
LMO7
|
LIM domain 7 |
| chr4_+_164754116 | 0.13 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr17_-_66220630 | 0.13 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr18_-_12656716 | 0.13 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr19_+_44212525 | 0.13 |
ENST00000391961.6
ENST00000621083.4 ENST00000313040.12 ENST00000586228.5 ENST00000588219.5 ENST00000589707.5 ENST00000588394.5 ENST00000589005.5 |
ZNF227
|
zinc finger protein 227 |
| chr11_-_26572130 | 0.13 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr3_+_159273235 | 0.13 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.4 | 1.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.7 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 1.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 3.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.5 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 2.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.7 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.3 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.4 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 1.0 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.5 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.5 | GO:0006067 | ethanol metabolic process(GO:0006067) |
| 0.0 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 1.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.5 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 1.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.8 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.0 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.5 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.9 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 1.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.7 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 2.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.8 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.0 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 3.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 1.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.8 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.2 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 2.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0043559 | insulin-like growth factor II binding(GO:0031995) insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |