Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for GATA1_GATA4
Z-value: 0.70
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
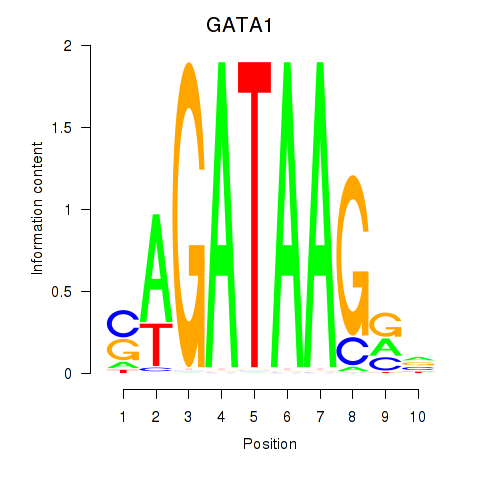
GATA1
|
ENSG00000102145.15 | GATA binding protein 1 |
|
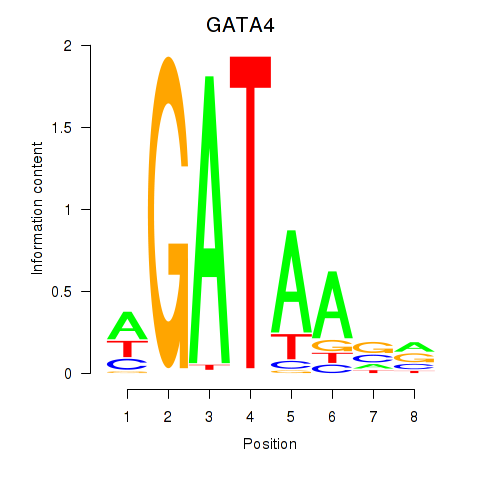
GATA4
|
ENSG00000136574.19 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA4 | hg38_v1_chr8_+_11704151_11704242, hg38_v1_chr8_+_11676952_11676966 | 0.33 | 4.2e-01 | Click! |
| GATA1 | hg38_v1_chrX_+_48786578_48786597 | 0.30 | 4.6e-01 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_22169580 | 1.00 |
ENST00000269216.10
|
GATA6
|
GATA binding protein 6 |
| chr10_-_127892930 | 0.68 |
ENST00000368671.4
|
CLRN3
|
clarin 3 |
| chr8_+_123182635 | 0.45 |
ENST00000276699.10
ENST00000522648.5 |
FAM83A
|
family with sequence similarity 83 member A |
| chr3_+_148865288 | 0.40 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3 |
| chr19_-_51019699 | 0.39 |
ENST00000358789.8
|
KLK10
|
kallikrein related peptidase 10 |
| chr2_+_196639686 | 0.37 |
ENST00000389175.9
|
CCDC150
|
coiled-coil domain containing 150 |
| chr1_-_24143112 | 0.32 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr6_+_12290353 | 0.32 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chr8_-_15238423 | 0.32 |
ENST00000382080.6
|
SGCZ
|
sarcoglycan zeta |
| chr11_-_116837586 | 0.32 |
ENST00000375320.5
ENST00000359492.6 ENST00000375329.6 ENST00000375323.5 ENST00000236850.5 |
APOA1
|
apolipoprotein A1 |
| chr4_-_102345196 | 0.30 |
ENST00000683412.1
ENST00000682227.1 |
SLC39A8
|
solute carrier family 39 member 8 |
| chr1_-_201399525 | 0.29 |
ENST00000367313.4
|
LAD1
|
ladinin 1 |
| chr6_+_1389553 | 0.29 |
ENST00000645481.2
|
FOXF2
|
forkhead box F2 |
| chr12_+_56083308 | 0.28 |
ENST00000683164.1
ENST00000415288.6 ENST00000683018.1 |
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr19_+_35248994 | 0.28 |
ENST00000427250.5
ENST00000605618.6 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr1_-_225999312 | 0.28 |
ENST00000272091.8
|
SDE2
|
SDE2 telomere maintenance homolog |
| chr1_-_201399302 | 0.28 |
ENST00000633953.1
ENST00000391967.7 |
LAD1
|
ladinin 1 |
| chr19_+_35248879 | 0.27 |
ENST00000347609.8
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr11_+_34621065 | 0.26 |
ENST00000257831.8
|
EHF
|
ETS homologous factor |
| chr1_-_209652329 | 0.26 |
ENST00000367030.7
ENST00000356082.9 |
LAMB3
|
laminin subunit beta 3 |
| chrX_-_79367307 | 0.25 |
ENST00000373298.7
|
ITM2A
|
integral membrane protein 2A |
| chr1_+_154321076 | 0.25 |
ENST00000324978.8
|
AQP10
|
aquaporin 10 |
| chr5_+_132073782 | 0.24 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2 |
| chr5_+_36608146 | 0.24 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr1_-_175192911 | 0.24 |
ENST00000444639.5
|
KIAA0040
|
KIAA0040 |
| chr1_+_34782259 | 0.23 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chrX_+_44844015 | 0.22 |
ENST00000339042.6
|
DUSP21
|
dual specificity phosphatase 21 |
| chr9_+_87726086 | 0.22 |
ENST00000677821.1
ENST00000677019.1 ENST00000678649.1 ENST00000342020.6 ENST00000677262.1 ENST00000343150.10 ENST00000677864.1 ENST00000340342.11 ENST00000676881.1 ENST00000676769.1 ENST00000678596.1 ENST00000679030.1 ENST00000678599.1 ENST00000677761.1 |
CTSL
|
cathepsin L |
| chr20_+_45306834 | 0.22 |
ENST00000343694.8
ENST00000372741.7 ENST00000372743.5 |
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region like |
| chr16_-_81220370 | 0.22 |
ENST00000337114.8
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chrX_+_47078380 | 0.21 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chrX_+_47078434 | 0.21 |
ENST00000397180.6
|
RGN
|
regucalcin |
| chr11_+_34621109 | 0.21 |
ENST00000450654.6
|
EHF
|
ETS homologous factor |
| chr1_+_206865620 | 0.21 |
ENST00000367098.6
|
IL20
|
interleukin 20 |
| chr3_-_98593589 | 0.21 |
ENST00000647941.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr1_+_154321107 | 0.20 |
ENST00000484864.1
|
AQP10
|
aquaporin 10 |
| chr1_+_202203721 | 0.20 |
ENST00000255432.11
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6 |
| chr6_-_11778781 | 0.20 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr11_-_124441158 | 0.20 |
ENST00000328064.2
|
OR8B8
|
olfactory receptor family 8 subfamily B member 8 |
| chr1_-_93585071 | 0.20 |
ENST00000539242.5
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr19_-_12887188 | 0.20 |
ENST00000264834.6
|
KLF1
|
Kruppel like factor 1 |
| chr17_+_76385256 | 0.20 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr8_-_20183090 | 0.20 |
ENST00000265808.11
ENST00000522513.5 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr1_-_225427897 | 0.20 |
ENST00000421383.1
ENST00000272163.9 |
LBR
|
lamin B receptor |
| chr14_+_64214136 | 0.19 |
ENST00000557084.1
ENST00000458046.6 |
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr12_-_120327762 | 0.19 |
ENST00000308366.9
ENST00000423423.3 |
PLA2G1B
|
phospholipase A2 group IB |
| chr2_+_147845020 | 0.19 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A |
| chr8_-_20183127 | 0.18 |
ENST00000276373.10
ENST00000519026.5 ENST00000440926.3 ENST00000437980.3 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr15_-_43220989 | 0.18 |
ENST00000540029.5
ENST00000441366.7 ENST00000648595.1 |
EPB42
|
erythrocyte membrane protein band 4.2 |
| chr3_-_119660580 | 0.18 |
ENST00000493094.6
ENST00000264231.7 ENST00000468801.1 |
POPDC2
|
popeye domain containing 2 |
| chr1_+_25272492 | 0.18 |
ENST00000454452.6
|
RHD
|
Rh blood group D antigen |
| chr5_+_67004618 | 0.17 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_188291854 | 0.17 |
ENST00000409830.6
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr7_+_150852507 | 0.17 |
ENST00000416793.6
ENST00000360937.9 ENST00000483043.1 |
AOC1
|
amine oxidase copper containing 1 |
| chr6_+_29306626 | 0.17 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr13_-_46182136 | 0.17 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr2_+_102311502 | 0.16 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr19_-_50983815 | 0.16 |
ENST00000391807.5
ENST00000593904.1 ENST00000595820.6 |
KLK7
|
kallikrein related peptidase 7 |
| chr12_+_8992029 | 0.16 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr18_+_63777773 | 0.16 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr8_+_32721823 | 0.16 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr5_+_140841183 | 0.16 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr11_-_125778788 | 0.16 |
ENST00000436890.2
|
PATE2
|
prostate and testis expressed 2 |
| chr6_+_137871208 | 0.15 |
ENST00000614035.4
ENST00000621150.3 ENST00000619035.4 ENST00000615468.4 ENST00000620204.3 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr1_+_200027605 | 0.15 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr1_-_177164673 | 0.15 |
ENST00000424564.2
ENST00000361833.7 |
ASTN1
|
astrotactin 1 |
| chrX_+_152914426 | 0.15 |
ENST00000318504.11
ENST00000449285.6 ENST00000539731.5 ENST00000535861.5 ENST00000370268.8 ENST00000370270.6 |
ZNF185
|
zinc finger protein 185 with LIM domain |
| chr1_-_43367689 | 0.15 |
ENST00000621943.4
|
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chrX_+_38352573 | 0.15 |
ENST00000039007.5
|
OTC
|
ornithine transcarbamylase |
| chr2_+_188291661 | 0.15 |
ENST00000409843.5
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr4_-_102345061 | 0.15 |
ENST00000394833.6
|
SLC39A8
|
solute carrier family 39 member 8 |
| chr12_-_70637405 | 0.15 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr12_+_8950036 | 0.14 |
ENST00000539240.5
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr10_+_88664439 | 0.14 |
ENST00000394375.7
ENST00000608620.5 ENST00000238983.9 ENST00000355843.2 |
LIPF
|
lipase F, gastric type |
| chr1_-_99766620 | 0.14 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr9_-_112175185 | 0.14 |
ENST00000355396.7
|
SUSD1
|
sushi domain containing 1 |
| chr6_+_125219804 | 0.14 |
ENST00000524679.1
|
TPD52L1
|
TPD52 like 1 |
| chrX_+_48521788 | 0.14 |
ENST00000651615.1
ENST00000495186.6 |
ENSG00000286268.1
EBP
|
novel protein EBP cholestenol delta-isomerase |
| chr4_-_71784046 | 0.14 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr11_-_59845496 | 0.14 |
ENST00000257248.3
|
CBLIF
|
cobalamin binding intrinsic factor |
| chr15_-_51738095 | 0.14 |
ENST00000560491.2
|
LYSMD2
|
LysM domain containing 2 |
| chr15_+_78340344 | 0.14 |
ENST00000299529.7
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr19_-_13102848 | 0.14 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr12_-_70754631 | 0.14 |
ENST00000440835.6
ENST00000549308.5 ENST00000550661.1 ENST00000378778.5 |
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr6_+_30647008 | 0.14 |
ENST00000293604.10
ENST00000376473.9 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr2_+_188291911 | 0.14 |
ENST00000410051.5
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr1_+_81306096 | 0.13 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr16_+_69105636 | 0.13 |
ENST00000569188.6
|
HAS3
|
hyaluronan synthase 3 |
| chr15_-_34336749 | 0.13 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr11_-_1036706 | 0.13 |
ENST00000421673.7
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr6_-_170291053 | 0.13 |
ENST00000366756.4
|
DLL1
|
delta like canonical Notch ligand 1 |
| chr10_+_80413817 | 0.13 |
ENST00000372187.9
|
PRXL2A
|
peroxiredoxin like 2A |
| chr19_-_35501878 | 0.13 |
ENST00000593342.5
ENST00000601650.1 ENST00000408915.6 |
DMKN
|
dermokine |
| chr17_-_31314040 | 0.13 |
ENST00000330927.5
|
EVI2B
|
ecotropic viral integration site 2B |
| chr10_-_94362925 | 0.13 |
ENST00000371361.3
|
NOC3L
|
NOC3 like DNA replication regulator |
| chr6_+_30647109 | 0.13 |
ENST00000651131.1
ENST00000376471.8 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr17_-_41124178 | 0.13 |
ENST00000394014.2
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr7_-_20217342 | 0.12 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr2_-_40512361 | 0.12 |
ENST00000403092.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr4_+_153466324 | 0.12 |
ENST00000409663.7
ENST00000409959.8 |
TMEM131L
|
transmembrane 131 like |
| chr3_+_189789643 | 0.12 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chr7_-_142962206 | 0.12 |
ENST00000460479.2
ENST00000476829.5 ENST00000355265.7 |
KEL
|
Kell metallo-endopeptidase (Kell blood group) |
| chr15_-_34318761 | 0.12 |
ENST00000290209.9
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr15_-_78944985 | 0.11 |
ENST00000615999.5
ENST00000677789.1 ENST00000676880.1 ENST00000677936.1 ENST00000220166.10 ENST00000677810.1 ENST00000678644.1 ENST00000677534.1 ENST00000677316.1 |
CTSH
|
cathepsin H |
| chr19_+_10286944 | 0.11 |
ENST00000380770.5
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr12_+_53268289 | 0.11 |
ENST00000257934.9
|
ESPL1
|
extra spindle pole bodies like 1, separase |
| chr1_+_14929734 | 0.11 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr19_+_10286971 | 0.11 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr19_-_50984028 | 0.11 |
ENST00000597707.5
|
KLK7
|
kallikrein related peptidase 7 |
| chr13_-_31162341 | 0.11 |
ENST00000445273.6
ENST00000630972.2 |
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chrX_-_15664798 | 0.11 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator |
| chr19_+_15640880 | 0.11 |
ENST00000586182.6
ENST00000221307.13 ENST00000591058.5 |
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chr1_-_32817311 | 0.11 |
ENST00000373477.9
ENST00000675785.1 |
YARS1
|
tyrosyl-tRNA synthetase 1 |
| chr6_+_130018565 | 0.11 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chrX_+_108044967 | 0.11 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_+_40988513 | 0.11 |
ENST00000649215.1
|
CTPS1
|
CTP synthase 1 |
| chr8_-_90082871 | 0.11 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr15_-_43879835 | 0.11 |
ENST00000636859.1
|
FRMD5
|
FERM domain containing 5 |
| chr9_-_6565462 | 0.10 |
ENST00000638661.1
ENST00000640208.1 |
GLDC
|
glycine decarboxylase |
| chr15_+_40844018 | 0.10 |
ENST00000344051.8
ENST00000562057.6 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr16_+_71526419 | 0.10 |
ENST00000539698.4
|
CHST4
|
carbohydrate sulfotransferase 4 |
| chrX_-_24647300 | 0.10 |
ENST00000379144.7
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr10_+_116591010 | 0.10 |
ENST00000530319.5
ENST00000527980.5 ENST00000471549.5 ENST00000534537.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr2_+_32165841 | 0.10 |
ENST00000357055.7
ENST00000435660.5 ENST00000440718.5 ENST00000379343.6 ENST00000282587.9 ENST00000406369.2 |
SLC30A6
|
solute carrier family 30 member 6 |
| chr1_-_246931892 | 0.10 |
ENST00000648844.2
|
AHCTF1
|
AT-hook containing transcription factor 1 |
| chr1_-_225428813 | 0.10 |
ENST00000338179.6
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr1_+_86993009 | 0.10 |
ENST00000370548.3
|
ENSG00000267561.2
|
novel protein |
| chrX_+_108045050 | 0.10 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr14_-_22815421 | 0.10 |
ENST00000674313.1
ENST00000555959.1 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr5_+_51383394 | 0.10 |
ENST00000230658.12
|
ISL1
|
ISL LIM homeobox 1 |
| chr3_-_79767987 | 0.10 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1 |
| chr4_+_48986268 | 0.10 |
ENST00000226432.9
|
CWH43
|
cell wall biogenesis 43 C-terminal homolog |
| chr2_-_40512423 | 0.10 |
ENST00000402441.5
ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 member A1 |
| chr12_+_103965863 | 0.10 |
ENST00000392872.8
ENST00000537100.5 |
TDG
|
thymine DNA glycosylase |
| chrX_+_135309480 | 0.10 |
ENST00000635820.1
|
ETDC
|
embryonic testis differentiation homolog C |
| chr9_-_112175283 | 0.09 |
ENST00000374270.8
ENST00000374263.7 |
SUSD1
|
sushi domain containing 1 |
| chr10_+_116590956 | 0.09 |
ENST00000358834.9
ENST00000528052.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr14_+_22497657 | 0.09 |
ENST00000390496.1
|
TRAJ41
|
T cell receptor alpha joining 41 |
| chr14_-_22815801 | 0.09 |
ENST00000397532.9
|
SLC7A7
|
solute carrier family 7 member 7 |
| chr17_-_49764123 | 0.09 |
ENST00000240364.7
ENST00000506156.1 |
FAM117A
|
family with sequence similarity 117 member A |
| chr1_-_111503622 | 0.09 |
ENST00000369716.9
ENST00000241356.5 |
TMIGD3
ADORA3
|
transmembrane and immunoglobulin domain containing 3 adenosine A3 receptor |
| chrX_+_129779930 | 0.09 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr2_+_188291994 | 0.09 |
ENST00000409927.5
ENST00000409805.5 |
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr6_-_47042306 | 0.09 |
ENST00000371253.7
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr6_+_47698574 | 0.09 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr14_+_85530163 | 0.09 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr3_+_169911566 | 0.09 |
ENST00000428432.6
ENST00000335556.7 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr14_-_22815856 | 0.09 |
ENST00000554758.1
ENST00000397528.8 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr12_+_12070932 | 0.09 |
ENST00000308721.9
|
BCL2L14
|
BCL2 like 14 |
| chr6_+_47698538 | 0.09 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chrX_+_48521817 | 0.09 |
ENST00000446158.5
ENST00000414061.1 |
EBP
|
EBP cholestenol delta-isomerase |
| chr7_-_99784175 | 0.09 |
ENST00000651514.1
ENST00000336411.7 ENST00000415003.1 ENST00000354593.6 |
CYP3A4
|
cytochrome P450 family 3 subfamily A member 4 |
| chr11_-_125778818 | 0.09 |
ENST00000358524.8
|
PATE2
|
prostate and testis expressed 2 |
| chr12_-_9999176 | 0.09 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr8_+_27771942 | 0.09 |
ENST00000523566.5
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr19_+_54724479 | 0.09 |
ENST00000291860.1
ENST00000400864.3 |
KIR3DL3
ENSG00000215765.3
|
killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 3 novel transcript |
| chr4_-_112636858 | 0.09 |
ENST00000503172.5
ENST00000505019.6 ENST00000309071.9 |
ZGRF1
|
zinc finger GRF-type containing 1 |
| chr10_-_102837406 | 0.09 |
ENST00000369887.4
ENST00000638272.1 ENST00000639393.1 ENST00000638971.1 ENST00000638190.1 |
CYP17A1
|
cytochrome P450 family 17 subfamily A member 1 |
| chr12_-_21775581 | 0.08 |
ENST00000537950.1
ENST00000665145.1 |
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr20_-_5010284 | 0.08 |
ENST00000379333.5
|
SLC23A2
|
solute carrier family 23 member 2 |
| chr13_-_46142834 | 0.08 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr9_-_124507382 | 0.08 |
ENST00000373588.9
ENST00000620110.4 |
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr14_+_21042352 | 0.08 |
ENST00000298690.5
|
RNASE7
|
ribonuclease A family member 7 |
| chr1_+_174877430 | 0.08 |
ENST00000392064.6
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr3_-_151278535 | 0.08 |
ENST00000309170.8
|
P2RY14
|
purinergic receptor P2Y14 |
| chr17_-_75844334 | 0.08 |
ENST00000592386.5
ENST00000412096.6 ENST00000586147.1 ENST00000207549.9 |
UNC13D
|
unc-13 homolog D |
| chr14_+_85530127 | 0.08 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_+_151437040 | 0.08 |
ENST00000520111.5
ENST00000520701.5 ENST00000429484.6 |
SLC36A1
|
solute carrier family 36 member 1 |
| chr18_+_44700796 | 0.08 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr3_-_155676363 | 0.08 |
ENST00000494598.5
|
PLCH1
|
phospholipase C eta 1 |
| chr11_+_118359572 | 0.08 |
ENST00000252108.8
ENST00000431736.6 |
UBE4A
|
ubiquitination factor E4A |
| chr11_-_75096876 | 0.08 |
ENST00000641541.1
ENST00000641593.1 ENST00000641504.1 ENST00000641931.1 ENST00000647690.1 |
OR2AT4
ENSG00000284722.2
|
olfactory receptor family 2 subfamily AT member 4 novel transcript |
| chr20_-_290364 | 0.08 |
ENST00000382369.9
|
C20orf96
|
chromosome 20 open reading frame 96 |
| chr17_-_66229380 | 0.08 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr7_+_123655857 | 0.08 |
ENST00000458573.3
ENST00000456238.2 |
LMOD2
|
leiomodin 2 |
| chr11_+_10305065 | 0.08 |
ENST00000534464.1
ENST00000278175.10 ENST00000530439.1 ENST00000524948.5 ENST00000528655.5 ENST00000526492.4 ENST00000525063.2 |
ADM
|
adrenomedullin |
| chr21_+_33543031 | 0.08 |
ENST00000356577.10
ENST00000381692.6 ENST00000300278.8 ENST00000381679.8 |
SON
|
SON DNA and RNA binding protein |
| chr5_+_148063971 | 0.07 |
ENST00000398454.5
ENST00000359874.7 ENST00000508733.5 ENST00000256084.8 |
SPINK5
|
serine peptidase inhibitor Kazal type 5 |
| chr22_+_21420636 | 0.07 |
ENST00000407598.2
|
HIC2
|
HIC ZBTB transcriptional repressor 2 |
| chr7_+_117014881 | 0.07 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_172750715 | 0.07 |
ENST00000392692.8
ENST00000417960.5 ENST00000428567.5 ENST00000366090.6 ENST00000426894.5 |
ECT2
|
epithelial cell transforming 2 |
| chrX_-_24672654 | 0.07 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr11_+_60699605 | 0.07 |
ENST00000300226.7
|
MS4A8
|
membrane spanning 4-domains A8 |
| chr18_+_58221535 | 0.07 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr2_+_173354820 | 0.07 |
ENST00000347703.7
ENST00000410101.7 ENST00000410019.3 ENST00000306721.8 |
CDCA7
|
cell division cycle associated 7 |
| chr14_+_22271921 | 0.07 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr7_-_99735093 | 0.07 |
ENST00000611620.4
ENST00000620220.6 ENST00000336374.4 |
CYP3A7-CYP3A51P
CYP3A7
|
CYP3A7-CYP3A51P readthrough cytochrome P450 family 3 subfamily A member 7 |
| chr6_-_46921926 | 0.07 |
ENST00000283296.12
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr4_-_64409444 | 0.07 |
ENST00000381210.8
ENST00000507440.5 |
TECRL
|
trans-2,3-enoyl-CoA reductase like |
| chr2_+_105744876 | 0.07 |
ENST00000233154.9
ENST00000451463.6 |
NCK2
|
NCK adaptor protein 2 |
| chr11_-_5324297 | 0.07 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2 |
| chr1_+_12464912 | 0.07 |
ENST00000543766.2
|
VPS13D
|
vacuolar protein sorting 13 homolog D |
| chr17_+_19411220 | 0.07 |
ENST00000461366.2
|
RNF112
|
ring finger protein 112 |
| chr2_-_162071183 | 0.07 |
ENST00000678668.1
|
DPP4
|
dipeptidyl peptidase 4 |
| chr12_-_52553139 | 0.07 |
ENST00000267119.6
|
KRT71
|
keratin 71 |
| chr17_+_7650916 | 0.07 |
ENST00000250111.9
|
ATP1B2
|
ATPase Na+/K+ transporting subunit beta 2 |
| chr7_-_44141074 | 0.07 |
ENST00000457314.5
ENST00000447951.1 ENST00000431007.1 |
MYL7
|
myosin light chain 7 |
| chr22_-_29388530 | 0.07 |
ENST00000357586.7
ENST00000432560.6 ENST00000405198.6 ENST00000317368.11 |
AP1B1
|
adaptor related protein complex 1 subunit beta 1 |
| chr15_+_74541200 | 0.07 |
ENST00000622429.1
ENST00000346246.10 |
ARID3B
|
AT-rich interaction domain 3B |
| chr1_-_167518521 | 0.07 |
ENST00000362089.10
|
CD247
|
CD247 molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.1 | 0.3 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.4 | GO:1903610 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.1 | 0.6 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.3 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.1 | 0.4 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.5 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.3 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.4 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0060913 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.2 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.0 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.2 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.0 | GO:0097545 | radial spoke(GO:0001534) axonemal outer doublet(GO:0097545) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.2 | GO:0047750 | C-8 sterol isomerase activity(GO:0000247) cholestenol delta-isomerase activity(GO:0047750) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.1 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |